Pandas Notebook 2, ATM350 Spring 2025 #
Motivating Science Questions:#
What was the daily temperature and precipitation at Albany last year?
What were the the days with the most precipitation?
Motivating Technical Question:#
How can we use Pandas to do some basic statistical analyses of our data?
We’ll start by repeating some of the same steps we did in the first Pandas notebook.#
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
sns.set()
year = 2024
file = f'/spare11/atm350/common/data/climo_alb_{year}.csv'
Display the first five lines of this file using Python’s built-in readline function
fileObj = open(file)
nLines = 5
for n in range(nLines):
line = fileObj.readline()
print(line)
DATE,MAX,MIN,AVG,DEP,PCP,SNW,DPTH,HDD,CDD
2024-01-01,36,25,30.5,4.2,0.00,0.0,0,34,0
2024-01-02,38,24,31.0,4.9,0.00,0.0,0,34,0
2024-01-03,40,32,36.0,10.1,0.00,0.0,0,29,0
2024-01-04,43,19,31.0,5.3,T,T,0,34,0
df = pd.read_csv(file, dtype='string')
nRows = df.shape[0]
print (f"Number of rows = {nRows}" )
nCols = df.shape[1]
print (f"Number of columns = {nCols}")
date = df['DATE']
date = pd.to_datetime(date,format="%Y-%m-%d")
maxT = df['MAX'].astype("float32")
minT = df['MIN'].astype("float32")
Number of rows = 366
Number of columns = 10
Let’s generate the final timeseries we made in our first Pandas notebook, with all the “bells and whistles” included.#
from matplotlib.dates import DateFormatter, AutoDateLocator,HourLocator,DayLocator,MonthLocator
fig, ax = plt.subplots(figsize=(15,10))
ax.plot (date, maxT, color='red',label = "Max T")
ax.plot (date, minT, color='blue', label = "Min T")
ax.set_title (f"ALB Year {year}")
ax.set_xlabel('Date')
ax.set_ylabel('Temperature (°F)' )
ax.xaxis.set_major_locator(MonthLocator(interval=1))
dateFmt = DateFormatter('%b %d')
ax.xaxis.set_major_formatter(dateFmt)
ax.legend (loc="best")
<matplotlib.legend.Legend at 0x14d967219400>
Read in precip data. This will be more challenging due to the presence of T(races).#
Let’s remind ourselves what the Dataframe looks like, paying particular attention to the daily precip column (PCP).
df
| DATE | MAX | MIN | AVG | DEP | PCP | SNW | DPTH | HDD | CDD | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2024-01-01 | 36 | 25 | 30.5 | 4.2 | 0.00 | 0.0 | 0 | 34 | 0 |
| 1 | 2024-01-02 | 38 | 24 | 31.0 | 4.9 | 0.00 | 0.0 | 0 | 34 | 0 |
| 2 | 2024-01-03 | 40 | 32 | 36.0 | 10.1 | 0.00 | 0.0 | 0 | 29 | 0 |
| 3 | 2024-01-04 | 43 | 19 | 31.0 | 5.3 | T | T | 0 | 34 | 0 |
| 4 | 2024-01-05 | 31 | 16 | 23.5 | -2.0 | T | T | 0 | 41 | 0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 361 | 2024-12-27 | 27 | 10 | 18.5 | -8.9 | 0.03 | 0.3 | 2 | 46 | 0 |
| 362 | 2024-12-28 | 35 | 22 | 28.5 | 1.3 | 0.04 | 0.0 | 2 | 36 | 0 |
| 363 | 2024-12-29 | 57 | 33 | 45.0 | 18.1 | 0.04 | 0.0 | 1 | 20 | 0 |
| 364 | 2024-12-30 | 58 | 44 | 51.0 | 24.3 | 0.17 | 0.0 | 0 | 14 | 0 |
| 365 | 2024-12-31 | 51 | 30 | 40.5 | 14.0 | T | 0.0 | 0 | 24 | 0 |
366 rows × 10 columns
DataSeries called precip and populate it with the requisite column from our Dataframe. Then print out its values.# %load /spare11/atm350/common/mar06/02a.py
precip = df['PCP']
precip
0 0.00
1 0.00
2 0.00
3 T
4 T
...
361 0.03
362 0.04
363 0.04
364 0.17
365 T
Name: PCP, Length: 366, dtype: string
The task now is to convert these values from strings to floating point values. Our task is more complicated due to the presence of strings that are clearly not numerical … such as “T” for trace.#
As we did in the first Pandas notebook with max temperatures greater than or equal to 90, create a subset of our Dataframe that consists only of those days where precip was a trace.#
traceDays = df[precip=='T']
traceDays
| DATE | MAX | MIN | AVG | DEP | PCP | SNW | DPTH | HDD | CDD | |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 | 2024-01-04 | 43 | 19 | 31.0 | 5.3 | T | T | 0 | 34 | 0 |
| 4 | 2024-01-05 | 31 | 16 | 23.5 | -2.0 | T | T | 0 | 41 | 0 |
| 10 | 2024-01-11 | 41 | 36 | 38.5 | 14.0 | T | 0.0 | 0 | 26 | 0 |
| 16 | 2024-01-17 | 22 | 9 | 15.5 | -8.3 | T | T | 4 | 49 | 0 |
| 17 | 2024-01-18 | 30 | 10 | 20.0 | -3.8 | T | T | 3 | 45 | 0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 353 | 2024-12-19 | 40 | 27 | 33.5 | 4.0 | T | 0.0 | 0 | 31 | 0 |
| 357 | 2024-12-23 | 24 | -5 | 9.5 | -18.9 | T | T | 2 | 55 | 0 |
| 359 | 2024-12-25 | 27 | 8 | 17.5 | -10.4 | T | T | 2 | 47 | 0 |
| 360 | 2024-12-26 | 20 | 5 | 12.5 | -15.1 | T | T | 2 | 52 | 0 |
| 365 | 2024-12-31 | 51 | 30 | 40.5 | 14.0 | T | 0.0 | 0 | 24 | 0 |
78 rows × 10 columns
traceDays.shape
(78, 10)
traceDays.shape[0]
78
shape attribute.# %load /spare11/atm350/common/mar06/02b.py
print (df[precip=='T'].shape)
numTraceDays = df[precip == 'T'].shape[0]
print (f"The total # of days in Albany in {year} that had a trace of precipitation was {numTraceDays}")
(78, 10)
The total # of days in Albany in 2024 that had a trace of precipitation was 78
Getting back to our task of converting precip amounts from strings to floating point numbers, one thing we could do is to create a new array and populate it via a loop, where we’d use an if-else logical test to check for Trace values and set the precip value to 0.00 for each day accordingly.#
There is a more efficient way to do this, though!#
We use the loc method of Pandas to find all elements of a DataSeries with a certain value, and then change that value to something else, all in the same line of code!#
In this case, let’s set all values of ‘T’ to ‘0.00’#
The line below is what we want! Before we execute it, let’s break it up into pieces.
df.loc[df['PCP'] =='T', ['PCP']] = '0.00'
First, create a Series of booleans corresponding to the specified condition.#
df['PCP'] == 'T'
0 False
1 False
2 False
3 True
4 True
...
361 False
362 False
363 False
364 False
365 True
Name: PCP, Length: 366, dtype: boolean
Next, build on that cell by using loc to display all rows that correspond to the condition being True.#
df.loc[df['PCP'] == 'T']
| DATE | MAX | MIN | AVG | DEP | PCP | SNW | DPTH | HDD | CDD | |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 | 2024-01-04 | 43 | 19 | 31.0 | 5.3 | T | T | 0 | 34 | 0 |
| 4 | 2024-01-05 | 31 | 16 | 23.5 | -2.0 | T | T | 0 | 41 | 0 |
| 10 | 2024-01-11 | 41 | 36 | 38.5 | 14.0 | T | 0.0 | 0 | 26 | 0 |
| 16 | 2024-01-17 | 22 | 9 | 15.5 | -8.3 | T | T | 4 | 49 | 0 |
| 17 | 2024-01-18 | 30 | 10 | 20.0 | -3.8 | T | T | 3 | 45 | 0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 353 | 2024-12-19 | 40 | 27 | 33.5 | 4.0 | T | 0.0 | 0 | 31 | 0 |
| 357 | 2024-12-23 | 24 | -5 | 9.5 | -18.9 | T | T | 2 | 55 | 0 |
| 359 | 2024-12-25 | 27 | 8 | 17.5 | -10.4 | T | T | 2 | 47 | 0 |
| 360 | 2024-12-26 | 20 | 5 | 12.5 | -15.1 | T | T | 2 | 52 | 0 |
| 365 | 2024-12-31 | 51 | 30 | 40.5 | 14.0 | T | 0.0 | 0 | 24 | 0 |
78 rows × 10 columns
Further build this line of code by only returning the column of interest.#
df.loc[df['PCP'] =='T', ['PCP']]
| PCP | |
|---|---|
| 3 | T |
| 4 | T |
| 10 | T |
| 16 | T |
| 17 | T |
| ... | ... |
| 353 | T |
| 357 | T |
| 359 | T |
| 360 | T |
| 365 | T |
78 rows × 1 columns
Finally, we have arrived at the full line of code! Take the column of interest, in this case precip only on those days where a trace was measured, and set its value to 0.00.#
df.loc[df['PCP'] =='T', ['PCP']] = '0.00'
df['PCP']
0 0.00
1 0.00
2 0.00
3 0.00
4 0.00
...
361 0.03
362 0.04
363 0.04
364 0.17
365 0.00
Name: PCP, Length: 366, dtype: string
This operation actually modifies the Dataframe in place … i.e., the individual cell values have changed, and henceforth in the notebook, the Dataframe will reflect these changed values. We can prove this by printing out a row from a date that we know had a trace amount.#
But first, how do we simply print a specific row from a dataframe? Since we know that Jan. 4 had a trace of precip, try this:#
jan04 = df['DATE'] == '2024-01-04'
jan04
0 False
1 False
2 False
3 True
4 False
...
361 False
362 False
363 False
364 False
365 False
Name: DATE, Length: 366, dtype: boolean
That produces a series of booleans; the one matching our condition is True. Now we can retrieve all the values for this date.#
df[jan04]
| DATE | MAX | MIN | AVG | DEP | PCP | SNW | DPTH | HDD | CDD | |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 | 2024-01-04 | 43 | 19 | 31.0 | 5.3 | 0.00 | T | 0 | 34 | 0 |
We see that the precip has now been set to 0.00.#
Having done this check, and thus re-set the values, let’s now convert this series into floating point values.#
precip = df['PCP'].astype("float32")
precip
0 0.00
1 0.00
2 0.00
3 0.00
4 0.00
...
361 0.03
362 0.04
363 0.04
364 0.17
365 0.00
Name: PCP, Length: 366, dtype: float32
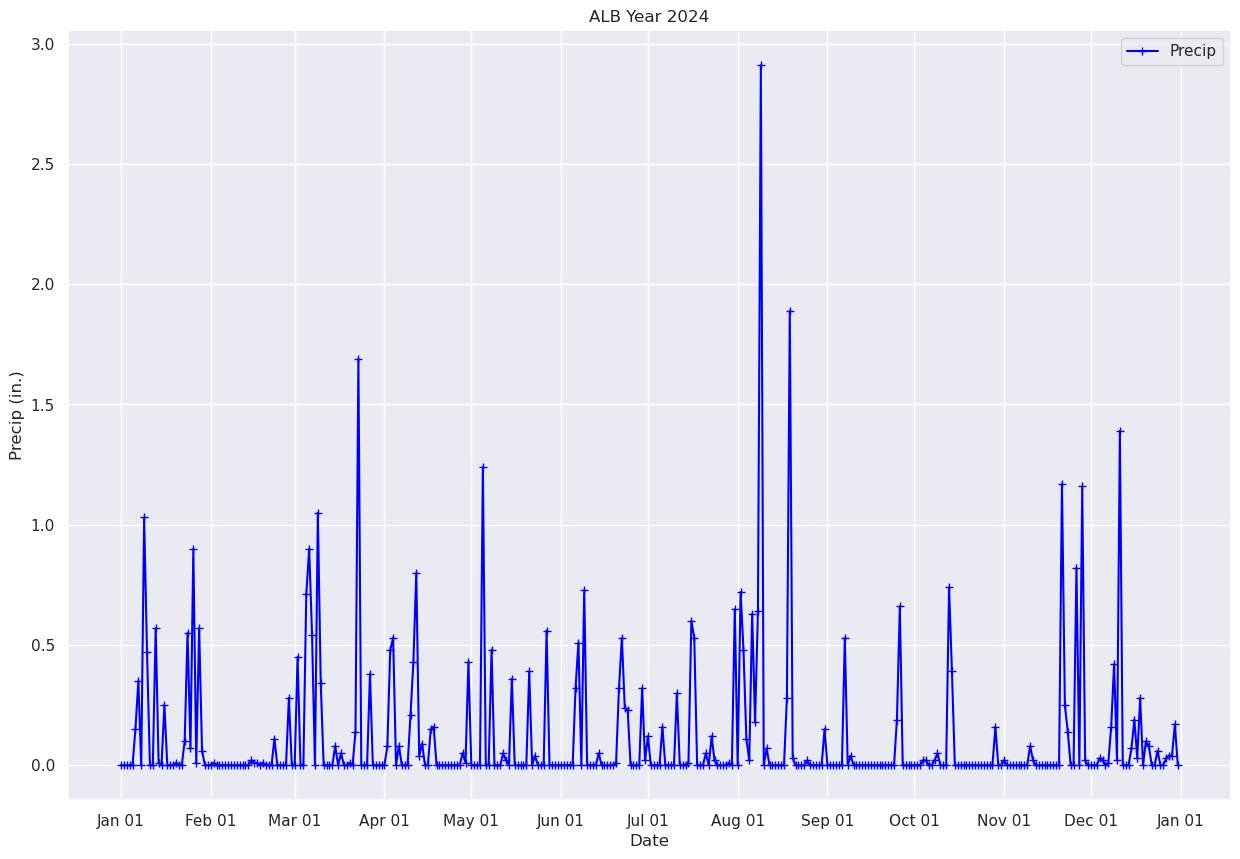
Plot each day’s precip total.#
fig, ax = plt.subplots(figsize=(15,10))
ax.plot (date, precip, color='blue', marker='+',label = "Precip")
ax.set_title (f"ALB Year {year}")
ax.set_xlabel('Date')
ax.set_ylabel('Precip (in.)' )
ax.xaxis.set_major_locator(MonthLocator(interval=1))
dateFmt = DateFormatter('%b %d')
ax.xaxis.set_major_formatter(dateFmt)
ax.legend (loc="best")
<matplotlib.legend.Legend at 0x14d966ff0500>
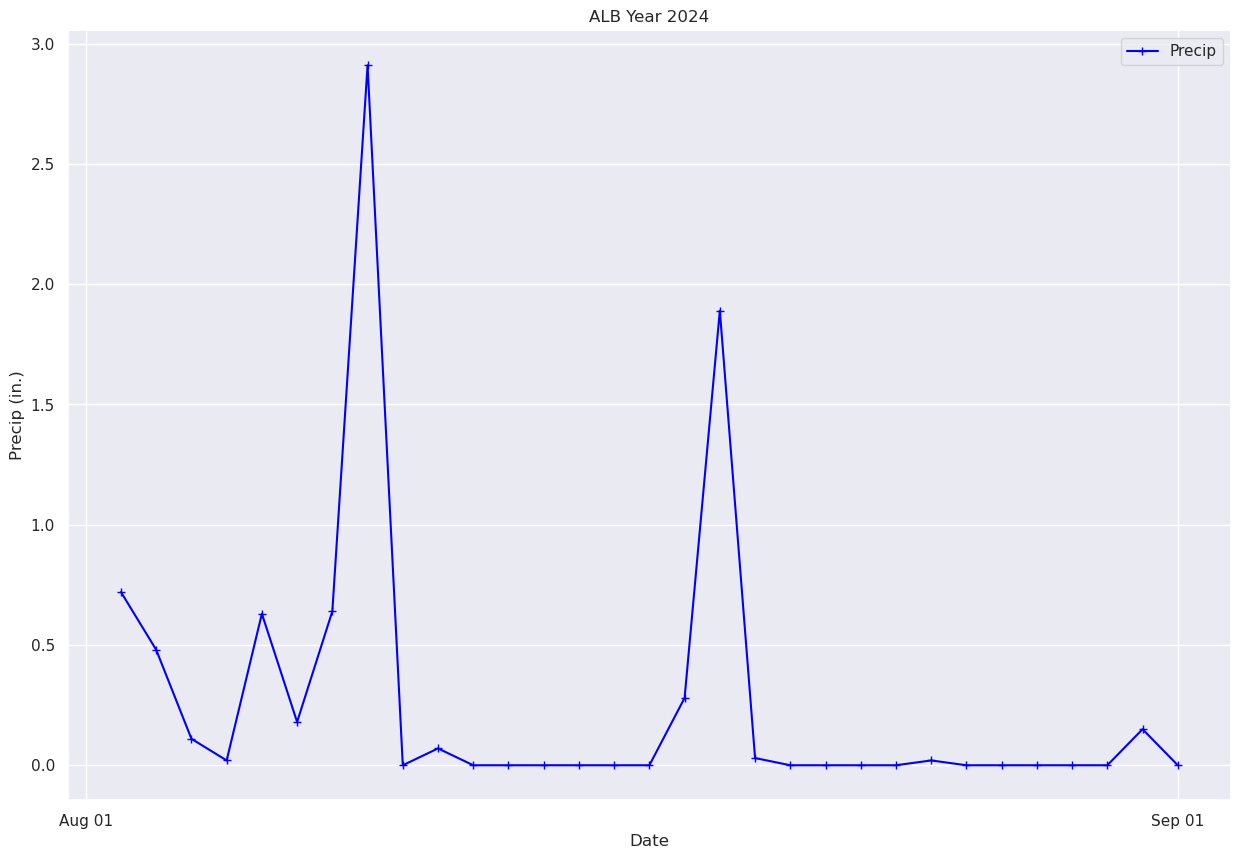
What if we just want to pick a certain time range? One simple way is to just pass in a subset of our x and y to the plot method.#
# Plot out just the trace for October. Corresponds to Julian days 215-246 ... thus, indices 214-245 (why?).
fig, ax = plt.subplots(figsize=(15,10))
ax.plot (date[214:245], precip[214:245], color='blue', marker='+',label = "Precip")
ax.set_title (f"ALB Year {year}")
ax.set_xlabel('Date')
ax.set_ylabel('Precip (in.)' )
ax.xaxis.set_major_locator(MonthLocator(interval=1))
dateFmt = DateFormatter('%b %d')
ax.xaxis.set_major_formatter(dateFmt)
ax.legend (loc="best")
<matplotlib.legend.Legend at 0x14d9670afe60>
# %load '/spare11/atm350/common/mar06/02c.py'
wetDays = df[precip>=1.00]
wetDays
| DATE | MAX | MIN | AVG | DEP | PCP | SNW | DPTH | HDD | CDD | |
|---|---|---|---|---|---|---|---|---|---|---|
| 8 | 2024-01-09 | 43 | 19 | 31.0 | 6.2 | 1.03 | 0.5 | 5 | 34 | 0 |
| 68 | 2024-03-09 | 45 | 36 | 40.5 | 7.3 | 1.05 | T | 0 | 24 | 0 |
| 82 | 2024-03-23 | 33 | 25 | 29.0 | -9.1 | 1.69 | 4.4 | 2 | 36 | 0 |
| 125 | 2024-05-05 | 57 | 49 | 53.0 | -3.2 | 1.24 | 0.0 | 0 | 12 | 0 |
| 221 | 2024-08-09 | 82 | 67 | 74.5 | 2.2 | 2.91 | 0.0 | 0 | 0 | 10 |
| 231 | 2024-08-19 | 82 | 59 | 70.5 | -0.7 | 1.89 | 0.0 | 0 | 0 | 6 |
| 325 | 2024-11-21 | 51 | 40 | 45.5 | 7.0 | 1.17 | 0.0 | 0 | 19 | 0 |
| 332 | 2024-11-28 | 39 | 33 | 36.0 | 0.0 | 1.16 | 3.8 | 0 | 29 | 0 |
| 345 | 2024-12-11 | 52 | 36 | 44.0 | 12.2 | 1.39 | T | 0 | 21 | 0 |
# Split the cell here so the table above will be displayed!
numWetDays = wetDays.shape[0]
print (f"The total # of days in Albany in {year} that had at least 1.00 in. of precip was {numWetDays}" )
The total # of days in Albany in 2024 that had at least 1.00 in. of precip was 9
Pandas has a function to compute the cumulative sum of a series. We’ll use it to compute and graph Albany’s total precip over the year.#
precipTotal = precip.cumsum()
precipTotal
0 0.000000
1 0.000000
2 0.000000
3 0.000000
4 0.000000
...
361 41.939995
362 41.979996
363 42.019997
364 42.189995
365 42.189995
Name: PCP, Length: 366, dtype: float32
We can see that the final total is in the last element of the precipTotal array. How can we explicitly print out just this value?#
One of the methods available to us in a Pandas DataSeries is values. Let’s display it:#
precipTotal.values
array([ 0. , 0. , 0. , 0. , 0. ,
0.15 , 0.5 , 0.5 , 1.53 , 2. ,
2. , 2. , 2.57 , 2.58 , 2.58 ,
2.83 , 2.83 , 2.83 , 2.83 , 2.84 ,
2.84 , 2.84 , 2.9399998, 3.4899998, 3.5599997,
4.4599996, 4.47 , 5.04 , 5.1 , 5.1 ,
5.1 , 5.1 , 5.11 , 5.11 , 5.11 ,
5.11 , 5.11 , 5.11 , 5.11 , 5.11 ,
5.11 , 5.11 , 5.11 , 5.11 , 5.11 ,
5.13 , 5.1400003, 5.1500006, 5.1500006, 5.160001 ,
5.160001 , 5.160001 , 5.160001 , 5.270001 , 5.270001 ,
5.270001 , 5.270001 , 5.270001 , 5.550001 , 5.550001 ,
5.550001 , 6.000001 , 6.000001 , 6.000001 , 6.710001 ,
7.610001 , 8.150002 , 8.150002 , 9.200002 , 9.540002 ,
9.540002 , 9.540002 , 9.540002 , 9.540002 , 9.620002 ,
9.620002 , 9.670002 , 9.670002 , 9.670002 , 9.680002 ,
9.680002 , 9.820003 , 11.510002 , 11.510002 , 11.510002 ,
11.510002 , 11.890002 , 11.890002 , 11.890002 , 11.890002 ,
11.890002 , 11.890002 , 11.970002 , 12.450002 , 12.980001 ,
12.980001 , 13.060001 , 13.060001 , 13.060001 , 13.060001 ,
13.270001 , 13.700002 , 14.500002 , 14.540002 , 14.630002 ,
14.630002 , 14.630002 , 14.780002 , 14.9400015, 14.9400015,
14.9400015, 14.9400015, 14.9400015, 14.9400015, 14.9400015,
14.9400015, 14.9400015, 14.9400015, 14.990002 , 15.000002 ,
15.430002 , 15.430002 , 15.430002 , 15.430002 , 15.430002 ,
16.670002 , 16.670002 , 16.670002 , 17.150002 , 17.150002 ,
17.150002 , 17.150002 , 17.2 , 17.220001 , 17.220001 ,
17.580002 , 17.580002 , 17.580002 , 17.580002 , 17.580002 ,
17.580002 , 17.970001 , 17.970001 , 18.010002 , 18.010002 ,
18.010002 , 18.010002 , 18.570002 , 18.570002 , 18.570002 ,
18.570002 , 18.570002 , 18.570002 , 18.570002 , 18.570002 ,
18.570002 , 18.570002 , 18.890001 , 19.400002 , 19.400002 ,
20.130001 , 20.130001 , 20.130001 , 20.130001 , 20.130001 ,
20.18 , 20.18 , 20.18 , 20.18 , 20.18 ,
20.18 , 20.19 , 20.51 , 21.04 , 21.28 ,
21.51 , 21.51 , 21.51 , 21.51 , 21.51 ,
21.83 , 21.85 , 21.970001 , 21.970001 , 21.970001 ,
21.970001 , 21.970001 , 22.130001 , 22.130001 , 22.130001 ,
22.130001 , 22.130001 , 22.43 , 22.43 , 22.43 ,
22.43 , 22.44 , 23.04 , 23.570002 , 23.570002 ,
23.570002 , 23.570002 , 23.62 , 23.62 , 23.740002 ,
23.760002 , 23.760002 , 23.760002 , 23.760002 , 23.760002 ,
23.770002 , 23.770002 , 24.420002 , 24.420002 , 25.140001 ,
25.62 , 25.730001 , 25.750002 , 26.380001 , 26.560001 ,
27.2 , 30.11 , 30.11 , 30.18 , 30.18 ,
30.18 , 30.18 , 30.18 , 30.18 , 30.18 ,
30.460001 , 32.350002 , 32.38 , 32.38 , 32.38 ,
32.38 , 32.38 , 32.4 , 32.4 , 32.4 ,
32.4 , 32.4 , 32.4 , 32.550003 , 32.550003 ,
32.550003 , 32.550003 , 32.550003 , 32.550003 , 32.550003 ,
33.08 , 33.08 , 33.120003 , 33.120003 , 33.120003 ,
33.120003 , 33.120003 , 33.120003 , 33.120003 , 33.120003 ,
33.120003 , 33.120003 , 33.120003 , 33.120003 , 33.120003 ,
33.120003 , 33.120003 , 33.120003 , 33.31 , 33.97 ,
33.97 , 33.97 , 33.97 , 33.97 , 33.97 ,
33.97 , 33.97 , 33.99 , 34.010002 , 34.010002 ,
34.010002 , 34.030003 , 34.08 , 34.08 , 34.08 ,
34.08 , 34.820004 , 35.210003 , 35.210003 , 35.210003 ,
35.210003 , 35.210003 , 35.210003 , 35.210003 , 35.210003 ,
35.210003 , 35.210003 , 35.210003 , 35.210003 , 35.210003 ,
35.210003 , 35.210003 , 35.370003 , 35.370003 , 35.370003 ,
35.390003 , 35.390003 , 35.390003 , 35.390003 , 35.390003 ,
35.390003 , 35.390003 , 35.390003 , 35.390003 , 35.470005 ,
35.490005 , 35.490005 , 35.490005 , 35.490005 , 35.490005 ,
35.490005 , 35.490005 , 35.490005 , 35.490005 , 35.490005 ,
36.660004 , 36.910004 , 37.050003 , 37.050003 , 37.050003 ,
37.870003 , 37.870003 , 39.030003 , 39.050003 , 39.050003 ,
39.050003 , 39.050003 , 39.050003 , 39.08 , 39.100002 ,
39.100002 , 39.11 , 39.27 , 39.69 , 39.71 ,
41.1 , 41.1 , 41.1 , 41.1 , 41.17 ,
41.359997 , 41.389996 , 41.669994 , 41.669994 , 41.769993 ,
41.849995 , 41.849995 , 41.849995 , 41.909996 , 41.909996 ,
41.909996 , 41.939995 , 41.979996 , 42.019997 , 42.189995 ,
42.189995 ], dtype=float32)
# %load '/spare11/atm350/common/mar06/02d.py'
# Print out the value of the last element in the array
precipTotal.values[-1]
np.float32(42.189995)
Plot the timeseries of the cumulative precip for Albany over the year.#
fig, ax = plt.subplots(figsize=(15,10))
ax.plot (date, precipTotal, color='blue', marker='.',label = "Precip")
ax.set_title (f"ALB Year {year}")
ax.set_xlabel('Date')
ax.set_ylabel('Precip (in.)' )
ax.xaxis.set_major_locator(MonthLocator(interval=1))
dateFmt = DateFormatter('%b %d')
ax.xaxis.set_major_formatter(dateFmt)
ax.legend (loc="best")
<matplotlib.legend.Legend at 0x14d966affe60>
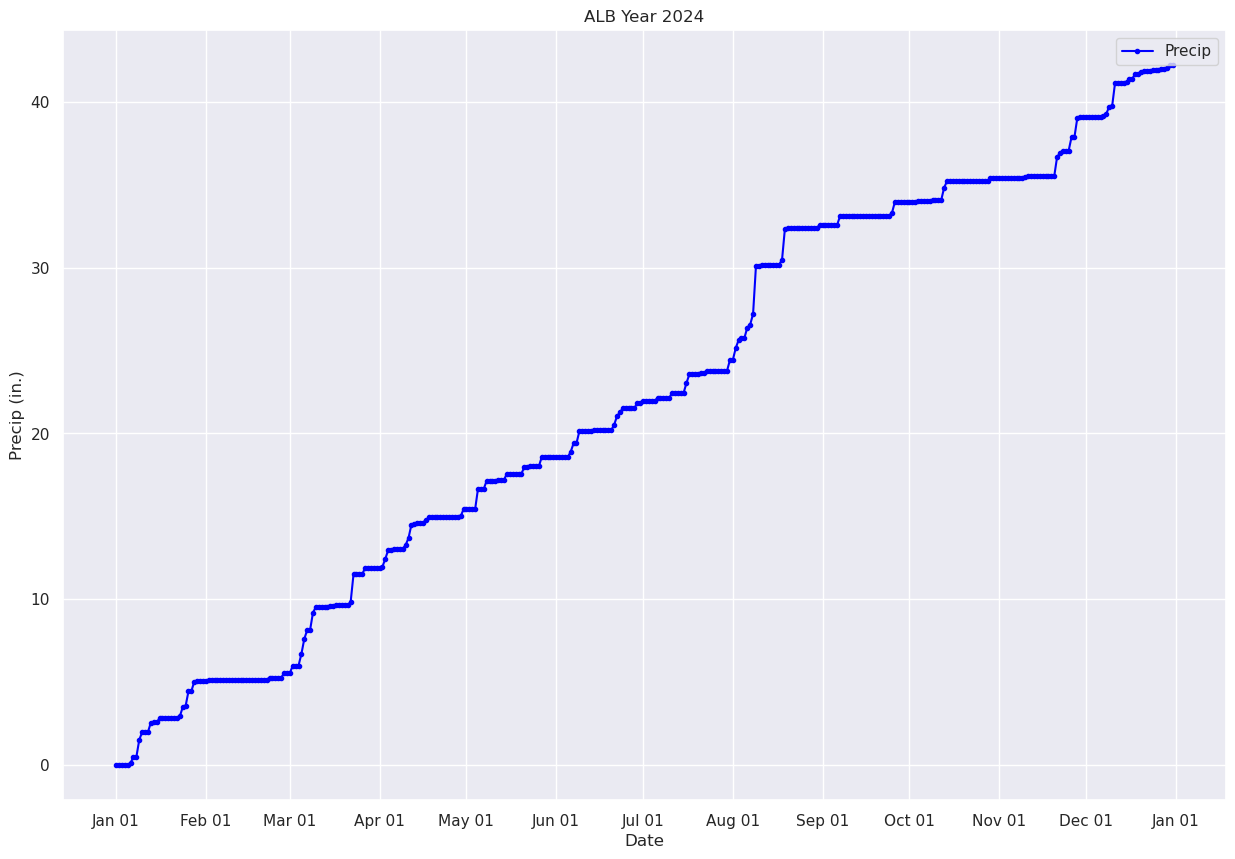
Pandas has a plethora of statistical analysis methods to apply on tabular data. An excellent summary method is describe.#
maxT.describe()
count 366.000000
mean 62.338799
std 19.533743
min 11.000000
25% 46.000000
50% 65.000000
75% 79.750000
max 96.000000
Name: MAX, dtype: float64
minT.describe()
count 366.000000
mean 42.967213
std 17.221647
min -5.000000
25% 30.000000
50% 41.000000
75% 56.750000
max 76.000000
Name: MIN, dtype: float64
precip.describe()
count 366.000000
mean 0.115273
std 0.297395
min 0.000000
25% 0.000000
50% 0.000000
75% 0.050000
max 2.910000
Name: PCP, dtype: float64
- First, express the condition where precip is equal to 0.00.
- Then, determine the # of rows of that resulting series.
# %load /spare11/atm350/common/mar06/02e.py
subset = precip[precip == 0.00]
nRows = subset.shape[0]
print (f"The number of days where precip was a trace or less was {nRows}")
print ("Since this is represents more than half the days of the years, the median must = 0")
The number of days where precip was a trace or less was 236
Since this is represents more than half the days of the years, the median must = 0
We’ll wrap up by calculating and then plotting rolling means over a period of days in the year, in order to smooth out the day-to-day variations.#
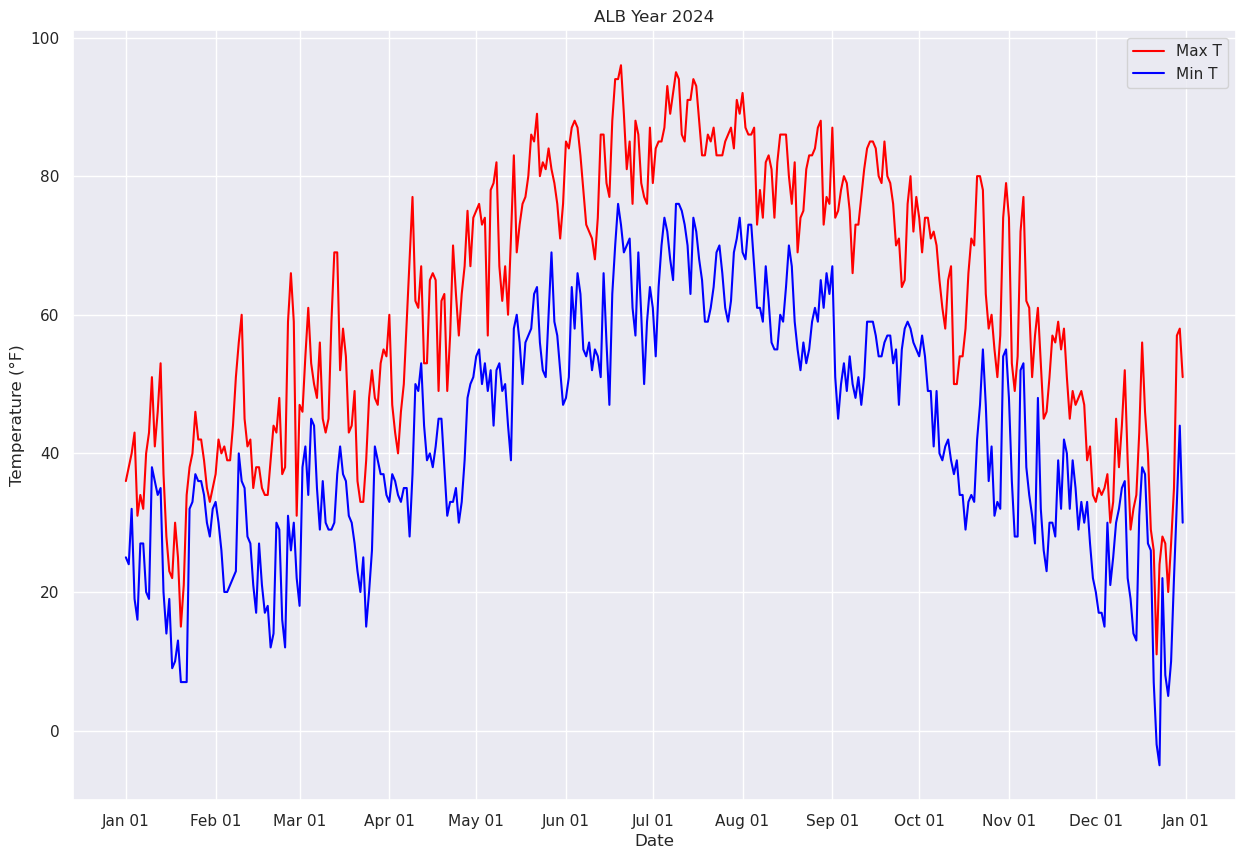
First, let’s replot the max and min temperature trace for the entire year, day-by-day.
fig, ax = plt.subplots(figsize=(15,10))
ax.plot (date, maxT, color='red',label = "Max T")
ax.plot (date, minT, color='blue', label = "Min T")
ax.set_title (f"ALB Year {year}")
ax.set_xlabel('Date')
ax.set_ylabel('Temperature (°F)' )
ax.xaxis.set_major_locator(MonthLocator(interval=1))
dateFmt = DateFormatter('%b %d')
ax.xaxis.set_major_formatter(dateFmt)
ax.legend (loc="best")
<matplotlib.legend.Legend at 0x14d966bb4500>

Now, let’s calculate and plot the daily mean temperature.
meanT = (maxT + minT) / 2.
meanT
0 30.5
1 31.0
2 36.0
3 31.0
4 23.5
...
361 18.5
362 28.5
363 45.0
364 51.0
365 40.5
Length: 366, dtype: float32
fig, ax = plt.subplots(figsize=(15,10))
ax.plot (date, meanT, color='green',label = "Mean T")
ax.set_title (f"ALB Year {year}")
ax.set_xlabel('Date')
ax.set_ylabel('Temperature (°F)' )
ax.xaxis.set_major_locator(MonthLocator(interval=1))
dateFmt = DateFormatter('%b %d')
ax.xaxis.set_major_formatter(dateFmt)
ax.legend (loc="best")
<matplotlib.legend.Legend at 0x14d9670382c0>
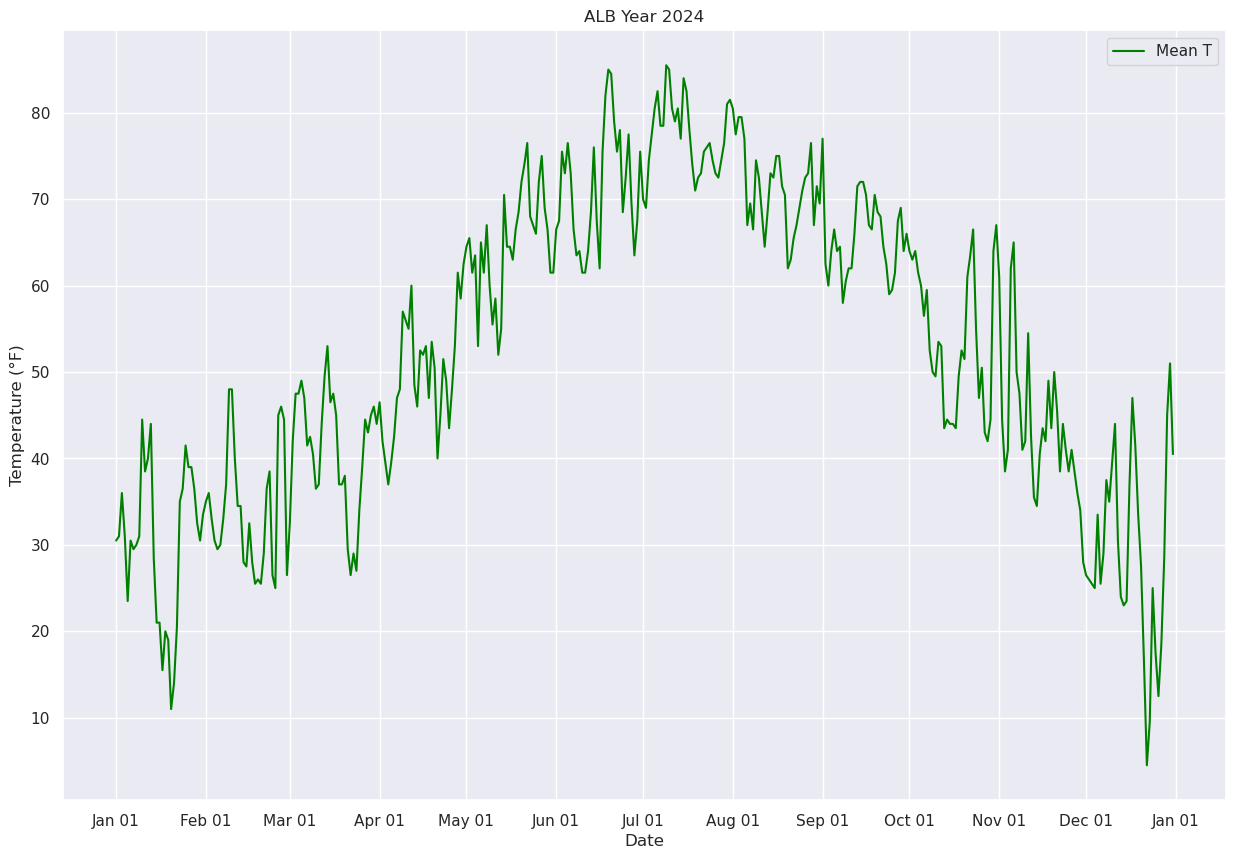
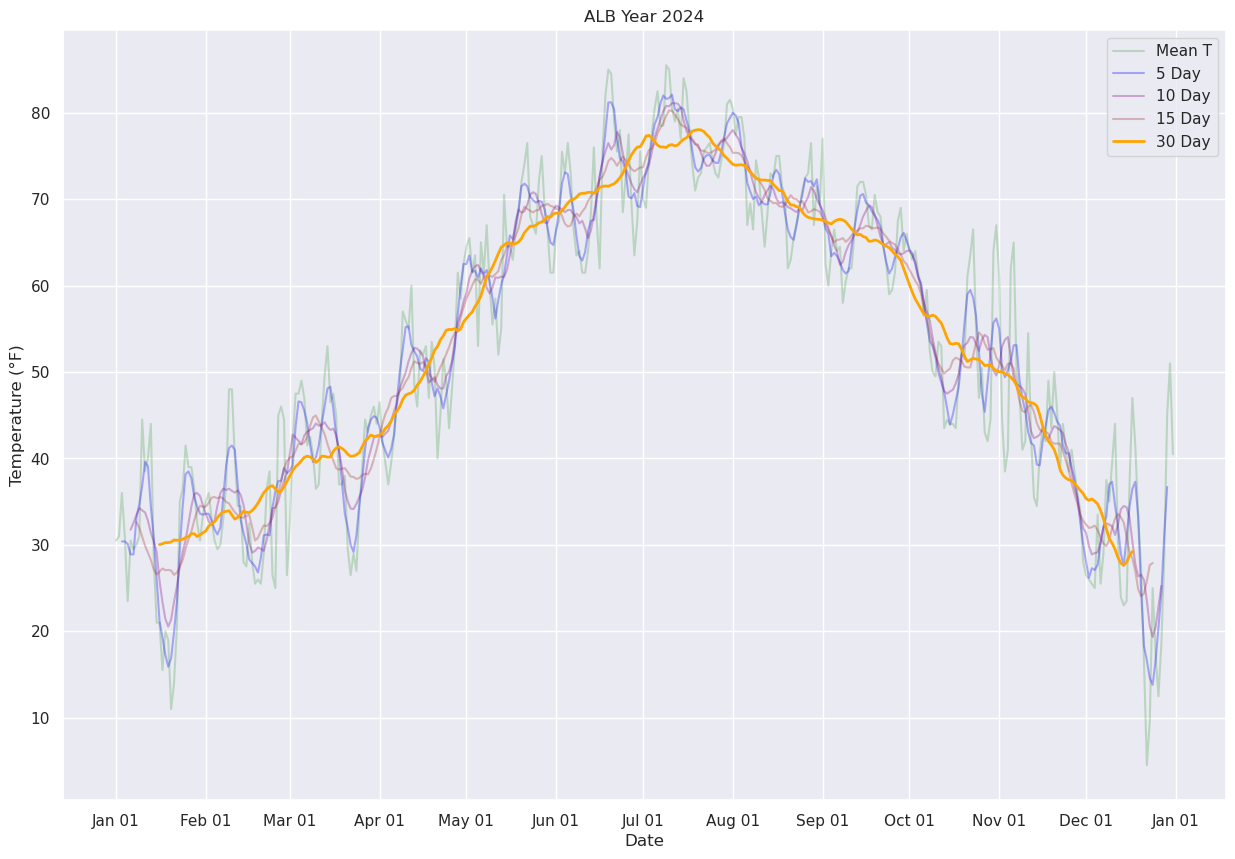
Next, let’s use Pandas’ rolling method to calculate the mean over a specified number of days. We’ll center the window at the midpoint of each period (thus, for a 30-day window, the first plotted point will be on Jan. 16 … covering the Jan. 1 –> Jan. 30 timeframe.
meanTr5 = meanT.rolling(window=5, center=True)
meanTr10 = meanT.rolling(window=10, center=True)
meanTr15 = meanT.rolling(window=15, center=True)
meanTr30 = meanT.rolling(window=30, center=True)
meanTr30.mean()
0 NaN
1 NaN
2 NaN
3 NaN
4 NaN
..
361 NaN
362 NaN
363 NaN
364 NaN
365 NaN
Length: 366, dtype: float64
meanTr5.mean()
0 NaN
1 NaN
2 30.4
3 30.4
4 30.1
...
361 24.4
362 31.1
363 36.7
364 NaN
365 NaN
Length: 366, dtype: float64
fig, ax = plt.subplots(figsize=(15,10))
ax.plot (date, meanT, color='green',label = "Mean T",alpha=0.2)
ax.plot (date, meanTr5.mean(), color='blue',label = "5 Day", alpha=0.3)
ax.plot (date, meanTr10.mean(), color='purple',label = "10 Day", alpha=0.3)
ax.plot (date, meanTr15.mean(), color='brown',label = "15 Day", alpha=0.3)
ax.plot (date, meanTr30.mean(), color='orange',label = "30 Day", alpha=1.0, linewidth=2)
ax.set_title (f"ALB Year {year}")
ax.set_xlabel('Date')
ax.set_ylabel('Temperature (°F)' )
ax.xaxis.set_major_locator(MonthLocator(interval=1))
dateFmt = DateFormatter('%b %d')
ax.xaxis.set_major_formatter(dateFmt)
ax.legend (loc="best")
<matplotlib.legend.Legend at 0x14d95e37fe60>
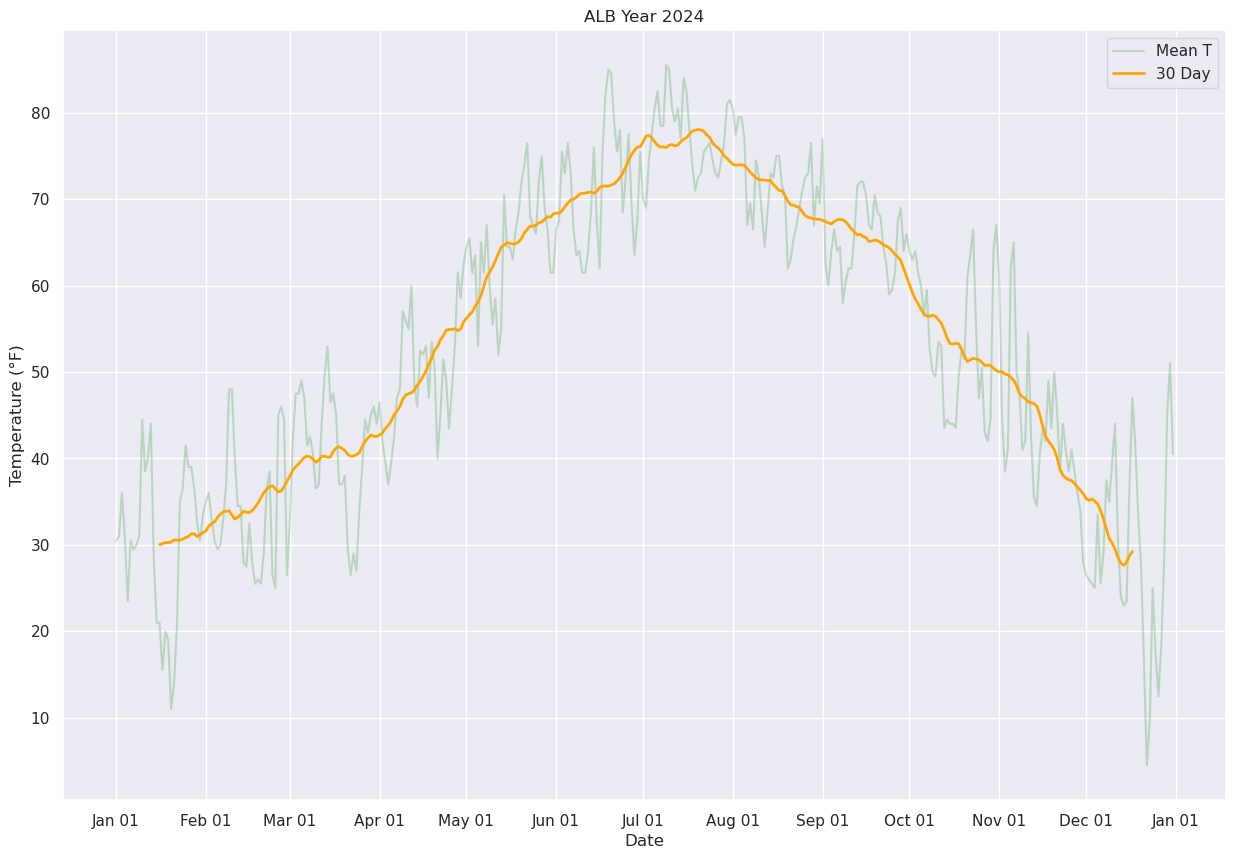
Display just the daily and 30-day running mean.
fig, ax = plt.subplots(figsize=(15,10))
ax.plot (date, meanT, color='green',label = "Mean T",alpha=0.2)
ax.plot (date, meanTr30.mean(), color='orange',label = "30 Day", alpha=1.0, linewidth=2)
ax.set_title (f"ALB Year {year}")
ax.set_xlabel('Date')
ax.set_ylabel('Temperature (°F)' )
ax.xaxis.set_major_locator(MonthLocator(interval=1))
dateFmt = DateFormatter('%b %d')
ax.xaxis.set_major_formatter(dateFmt)
ax.legend (loc="best")
<matplotlib.legend.Legend at 0x14d95e2b78f0>