ERA-5 End-to-End Data and Graphics Preparation for the Science-on-a-Sphere
Contents
ERA-5 End-to-End Data and Graphics Preparation for the Science-on-a-Sphere¶
This notebook will create all the necessary files and directories for a visualization on NOAA’s Science-on-a-Sphere. For this example, we will produce a map of 500 hPa geopotential heights and sea-level pressure, using ERA-5 reanalysis, for the January 26 1978 intense surface cyclone, aka The Cleveland Superbomb.
Overview:¶
Set up output directories for SoS
Programmatically request a specific date from the RDA ERA-5 THREDDS repository
Re-arrange the order of the longitudinal dimension in the dataset
Use Cartopy’s
add_cyclic_pointmethod to avoid a blank seam at the datelineCreate small and large thumbnail graphics.
Create a standalone colorbar
Create a set of labels for each plot
Create 24 hours worth of Science-on-a-Sphere-ready plots
Create an SoS playlist file
Prerequisites¶
Concepts |
Importance |
Notes |
|---|---|---|
Xarray |
Necessary |
|
Linux command line / directory structure |
Helpful |
Time to learn: 30 minutes
Imports¶
import xarray as xr
import numpy as np
import pandas as pd
from pandas.tseries.offsets import MonthEnd,MonthBegin
import cartopy.feature as cfeature
import cartopy.crs as ccrs
import cartopy.util as cutil
import requests
from datetime import datetime, timedelta
import warnings
import metpy.calc as mpcalc
from metpy.units import units
import matplotlib.pyplot as plt
ERROR 1: PROJ: proj_create_from_database: Open of /knight/anaconda_aug22/envs/aug22_env/share/proj failed
Do not output warning messages
warnings.simplefilter("ignore")
Set up output directories for SoS¶
The software that serves output for the Science-on-a-Sphere expects a directory structure as follows:
Top level directory: choose a name that is consistent with your case, e.g.: cle_superbomb
2nd level directory: choose a name that is consistent with the graphic, e.g.: SLP_500Z
3rd level directories:
2048: Contains the graphics (resolution: 2048x1024) that this notebook generates
labels: Contains one or two files:
(required) A text file,
labels.txt, which has as many lines as there are graphics in the 2048 file. Each line functions as the title of each graphic.(optional) A PNG file,
colorbar.png, which colorbar which will get overlaid on your map graphic.
media: Contains large and small thumbnails (
thumbnail_small.jpg,thumbnail_large.jpg) that serve as icons on the SoS iPad and SoS computer appsplaylist: A text file,
playlist.sos, which tells the SoS how to display your product
As an example, here is how the directory structure on our SoS looks for the products generated by this notebook. Our SoS computer stores locally-produced content in the /shared/sos/media/site-custom directory (note: the SoS directories are not network-accessible, so you won’t be able to cd into them). The ERA-5 visualizations go in the ERA5 subfolder. Your top-level directory sits within the ERA5 folder.
sos@sos1:/shared/sos/media/site-custom/ERA5/cle_superbomb/SLP_500Z$ ls -R
.:
2048 labels media playlist
./2048:
ERA5_1978012600-fs8.png ERA5_1978012606-fs8.png ERA5_1978012612-fs8.png ERA5_1978012618-fs8.png
ERA5_1978012601-fs8.png ERA5_1978012607-fs8.png ERA5_1978012613-fs8.png ERA5_1978012619-fs8.png
ERA5_1978012602-fs8.png ERA5_1978012608-fs8.png ERA5_1978012614-fs8.png ERA5_1978012620-fs8.png
ERA5_1978012603-fs8.png ERA5_1978012609-fs8.png ERA5_1978012615-fs8.png ERA5_1978012621-fs8.png
ERA5_1978012604-fs8.png ERA5_1978012610-fs8.png ERA5_1978012616-fs8.png ERA5_1978012622-fs8.png
ERA5_1978012605-fs8.png ERA5_1978012611-fs8.png ERA5_1978012617-fs8.png ERA5_1978012623-fs8.png
./labels:
colorbar.png labels.txt
./media:
thumbnail_big.jpg thumbnail_small.jpg
./playlist:
playlist.sos
Define the 1st and 2nd-level directories.
# You define these
caseDir = 'cle_superbomb'
prodDir = 'SLP_500Z'
The 3rd-level directories follow from the 1st and 2nd.
# These remain as is
graphicsDir = caseDir + '/' + prodDir + '/2048/'
labelsDir = caseDir + '/' + prodDir + '/labels/'
mediaDir = caseDir + '/' + prodDir + '/media/'
playlistDir = caseDir + '/' + prodDir + '/playlist/'
Create these directories via a Linux command
! mkdir -p {graphicsDir} {labelsDir} {mediaDir} {playlistDir}
! magic indicates that what follows is a Linux command.The
-p option for mkdir will create all subdirectories, and also will do nothing if the directories already exist.Programmatically request a specific date from the RDA ERA-5 THREDDS repository¶
The next several cells set the date/time, creates the URL strings, and retrieves selected grids from RDA.
# Select your date and time here
year = 1978
month = 1
day = 26
hour = 0
While Xarray can create datasets and data arrays from multiple files, via its
open_mfdataset method, this method does not reliably work when reading from a THREDDS server such as RDA's. Therefore, please restrict your temporal range to no more than one calendar day in this notebook.
validTime = datetime(year, month, day, hour)
YYYYMM = validTime.strftime("%Y%m")
YYYYMMDD = validTime.strftime("%Y%m%d")
Among the many tools for time series analysis that Pandas provides are functions to determine the first and last days of a month.
monthEnd = pd.to_datetime(YYYYMM, format='%Y%m') + MonthEnd(0)
mEndStr = monthEnd.strftime('%Y%m%d23')
monthBeg = pd.to_datetime(YYYYMM, format='%Y%m') + MonthBegin(0)
mBegStr = monthBeg.strftime('%Y%m%d00')
dayBegStr = YYYYMMDD + '00'
dayEndStr = YYYYMMDD + '23'
dayBeg = datetime.strptime(dayBegStr, '%Y%m%d%H')
dayEnd = datetime.strptime(dayEndStr, '%Y%m%d%H')
dayBeg, dayEnd
(datetime.datetime(1978, 1, 26, 0, 0), datetime.datetime(1978, 1, 26, 23, 0))
monthBeg, monthEnd
(Timestamp('1978-01-01 00:00:00'), Timestamp('1978-01-31 00:00:00'))
Create the URLs for the various grids.
SfcURL = []
SfcURL.append('https://rda.ucar.edu/thredds/dodsC/files/g/ds633.0/e5.oper.an.')
SfcURL.append(YYYYMM + '/e5.oper.an')
SfcURL.append('.ll025')
SfcURL.append('.' + mBegStr + '_' + mEndStr + '.nc')
print(SfcURL)
PrsURL = []
PrsURL.append('https://rda.ucar.edu/thredds/dodsC/files/g/ds633.0/e5.oper.an.')
PrsURL.append(YYYYMM + '/e5.oper.an')
PrsURL.append('.ll025')
PrsURL.append('.' + dayBegStr + '_' + dayEndStr + '.nc' )
print(PrsURL)
ds_urlZ = PrsURL[0] + 'pl/' + PrsURL[1] + '.pl.128_129_z' + PrsURL[2] + 'sc' + PrsURL[3]
ds_urlQ = PrsURL[0] + 'pl/' + PrsURL[1] + '.pl.128_133_q' + PrsURL[2] + 'sc' + PrsURL[3]
ds_urlT = PrsURL[0] + 'pl/' + PrsURL[1] + '.pl.128_130_t' + PrsURL[2] + 'sc' + PrsURL[3]
# Notice that U and V use "uv" instead of "sc" in their name!
ds_urlU = PrsURL[0] + 'pl/' + PrsURL[1] + '.pl.128_131_u' + PrsURL[2] + 'uv' + PrsURL[3]
ds_urlV = PrsURL[0] + 'pl/' + PrsURL[1] + '.pl.128_132_v' + PrsURL[2] + 'uv' + PrsURL[3]
ds_urlU10 = SfcURL[0] + 'sfc/' + SfcURL[1] + '.sfc.128_165_10u' + SfcURL[2] + 'sc' + SfcURL[3]
ds_urlV10 = SfcURL[0] + 'sfc/' + SfcURL[1] + '.sfc.128_166_10v' + SfcURL[2] + 'sc' + SfcURL[3]
ds_urlSLP = SfcURL[0] + 'sfc/' + SfcURL[1] + '.sfc.128_151_msl' + SfcURL[2] + 'sc' + SfcURL[3]
['https://rda.ucar.edu/thredds/dodsC/files/g/ds633.0/e5.oper.an.', '197801/e5.oper.an', '.ll025', '.1978010100_1978013123.nc']
['https://rda.ucar.edu/thredds/dodsC/files/g/ds633.0/e5.oper.an.', '197801/e5.oper.an', '.ll025', '.1978012600_1978012623.nc']
Use your RDA credentials to access these individual NetCDF files. You should have previously registered with RDA, and then saved your user id and password in your home directory as .rdarc , with permissions such that only you have read/write access.
#Retrieve login credential for RDA.
from pathlib import Path
HOME = str(Path.home())
credFile = open(HOME+'/.rdarc','r')
userId, pw = credFile.read().split()
Connect to the RDA THREDDS server and point to the files of interest. Here, we’ll comment out the ones we are not interested in but can always uncomment when we want them.
session = requests.Session()
session.auth = (userId, pw)
#storeU10 = xr.backends.PydapDataStore.open(ds_urlU10, session=session)
#storeV10 = xr.backends.PydapDataStore.open(ds_urlV10, session=session)
storeSLP = xr.backends.PydapDataStore.open(ds_urlSLP, session=session)
#storeQ = xr.backends.PydapDataStore.open(ds_urlQ, session=session)
#storeT = xr.backends.PydapDataStore.open(ds_urlT, session=session)
#storeU = xr.backends.PydapDataStore.open(ds_urlU, session=session)
#storeV = xr.backends.PydapDataStore.open(ds_urlV, session=session)
storeZ = xr.backends.PydapDataStore.open(ds_urlZ, session=session)
ds_urlSLP
'https://rda.ucar.edu/thredds/dodsC/files/g/ds633.0/e5.oper.an.sfc/197801/e5.oper.an.sfc.128_151_msl.ll025sc.1978010100_1978013123.nc'
#dsU10 = xr.open_dataset(storeU10)
#dsV10 = xr.open_dataset(storeV10)
dsSLP = xr.open_dataset(storeSLP)
#dsQ = xr.open_dataset(storeQ)
#dsT = xr.open_dataset(storeT)
#dsU = xr.open_dataset(storeU)
#dsV = xr.open_dataset(storeV)
dsZ = xr.open_dataset(storeZ)
Examine the datasets¶
The surface fields span the entire month. Let’s restrict the range so it matches the day of interest.
dsSLP = dsSLP.sel(time=slice(dayBeg,dayEnd))
Create an Xarray DataArray object for SLP and perform unit conversion
SLP = dsSLP['MSL']
SLP = SLP.metpy.convert_units('hPa')
Examine the SLP DataArray
SLP
<xarray.DataArray 'MSL' (time: 24, latitude: 721, longitude: 1440)>
<Quantity([[[1032.5399 1032.5399 1032.5399 ... 1032.5399 1032.5399 1032.5399 ]
[1032.82 1032.8225 1032.8225 ... 1032.8175 1032.8175 1032.82 ]
[1033.0275 1033.03 1033.0325 ... 1033.0225 1033.025 1033.025 ]
...
[1009.8475 1009.85 1009.85 ... 1009.83997 1009.84247 1009.845 ]
[1009.77747 1009.77747 1009.77496 ... 1009.77496 1009.77496 1009.77747]
[1009.69 1009.69 1009.69 ... 1009.69 1009.69 1009.69 ]]
[[1031.6238 1031.6238 1031.6238 ... 1031.6238 1031.6238 1031.6238 ]
[1031.9113 1031.9137 1031.9137 ... 1031.9087 1031.9113 1031.9113 ]
[1032.1162 1032.1188 1032.1212 ... 1032.1112 1032.1138 1032.1162 ]
...
[1009.84375 1009.84625 1009.84625 ... 1009.83875 1009.83875 1009.84125]
[1009.78625 1009.78625 1009.78625 ... 1009.78625 1009.78625 1009.78625]
[1009.70874 1009.70874 1009.70874 ... 1009.70874 1009.70874 1009.70874]]
[[1031.6018 1031.6018 1031.6018 ... 1031.6018 1031.6018 1031.6018 ]
[1031.8993 1031.8993 1031.8993 ... 1031.8944 1031.8969 1031.8969 ]
[1032.1194 1032.1218 1032.1244 ... 1032.1144 1032.1144 1032.1168 ]
...
...
...
[1006.08435 1006.08185 1006.07935 ... 1006.08936 1006.08435 1006.08435]
[1006.33185 1006.32935 1006.32684 ... 1006.33685 1006.33685 1006.33435]
[1006.89185 1006.89185 1006.89185 ... 1006.89185 1006.89185 1006.89185]]
[[1021.5062 1021.5062 1021.5062 ... 1021.5062 1021.5062 1021.5062 ]
[1021.96375 1021.96625 1021.96875 ... 1021.95624 1021.95874 1021.96124]
[1022.4337 1022.4387 1022.4437 ... 1022.4212 1022.4237 1022.4287 ]
...
[1009.1812 1009.1837 1009.1812 ... 1009.1787 1009.1787 1009.1787 ]
[1008.89374 1008.89124 1008.89124 ... 1008.89374 1008.89374 1008.89374]
[1008.97125 1008.97125 1008.97125 ... 1008.97125 1008.97125 1008.97125]]
[[1021.305 1021.305 1021.305 ... 1021.305 1021.305 1021.305 ]
[1021.72 1021.72 1021.7225 ... 1021.71246 1021.71497 1021.71747]
[1022.13995 1022.14496 1022.14996 ... 1022.13 1022.1325 1022.13745]
...
[1009.005 1009.0075 1009.00995 ... 1009. 1009. 1009. ]
[1008.6925 1008.6925 1008.6925 ... 1008.6925 1008.6925 1008.6925 ]
[1008.8075 1008.8075 1008.8075 ... 1008.8075 1008.8075 1008.8075 ]]], 'hectopascal')>
Coordinates:
* latitude (latitude) float64 90.0 89.75 89.5 89.25 ... -89.5 -89.75 -90.0
* longitude (longitude) float64 0.0 0.25 0.5 0.75 ... 359.0 359.2 359.5 359.8
* time (time) datetime64[ns] 1978-01-26 ... 1978-01-26T23:00:00
Attributes:
long_name: Mean sea level pressure
short_name: msl
original_format: WMO GRIB 1 with ECMWF...
ecmwf_local_table: 128
ecmwf_parameter: 151
minimum_value: 93279.19
maximum_value: 106472.31
grid_specification: 0.25 degree x 0.25 de...
rda_dataset: ds633.0
rda_dataset_url: https:/rda.ucar.edu/d...
rda_dataset_doi: DOI: 10.5065/BH6N-5N20
rda_dataset_group: ERA5 atmospheric surf...
QuantizeGranularBitGroomNumberOfSignificantDigits: 7
_ChunkSizes: [27, 139, 277]Now examine the geopotential data on isobaric surfaces.
dsZ
<xarray.Dataset>
Dimensions: (time: 24, level: 37, latitude: 721, longitude: 1440)
Coordinates:
* latitude (latitude) float64 90.0 89.75 89.5 89.25 ... -89.5 -89.75 -90.0
* level (level) float64 1.0 2.0 3.0 5.0 7.0 ... 925.0 950.0 975.0 1e+03
* longitude (longitude) float64 0.0 0.25 0.5 0.75 ... 359.0 359.2 359.5 359.8
* time (time) datetime64[ns] 1978-01-26 ... 1978-01-26T23:00:00
Data variables:
Z (time, level, latitude, longitude) float32 ...
utc_date (time) int32 1978012600 1978012601 ... 1978012622 1978012623
Attributes:
DATA_SOURCE: ECMWF: https://cds.climate.copernicus.eu...
NETCDF_CONVERSION: CISL RDA: Conversion from ECMWF GRIB 1 d...
NETCDF_VERSION: 4.8.1
CONVERSION_PLATFORM: Linux r1i3n32 4.12.14-95.51-default #1 S...
CONVERSION_DATE: Sun Jul 3 14:02:30 MDT 2022
Conventions: CF-1.6
NETCDF_COMPRESSION: NCO: Precision-preserving compression to...
history: Sun Jul 3 14:03:04 2022: ncks -4 --ppc ...
NCO: netCDF Operators version 5.0.3 (Homepage...
DODS_EXTRA.Unlimited_Dimension: timeTo reduce the memory footprint for data on isobaric surfaces, let’s request only 6 out of the 37 available levels.
dsZ = dsZ.sel(level = [200,300,500,700,850,1000])
dsZ
<xarray.Dataset>
Dimensions: (time: 24, level: 6, latitude: 721, longitude: 1440)
Coordinates:
* latitude (latitude) float64 90.0 89.75 89.5 89.25 ... -89.5 -89.75 -90.0
* level (level) float64 200.0 300.0 500.0 700.0 850.0 1e+03
* longitude (longitude) float64 0.0 0.25 0.5 0.75 ... 359.0 359.2 359.5 359.8
* time (time) datetime64[ns] 1978-01-26 ... 1978-01-26T23:00:00
Data variables:
Z (time, level, latitude, longitude) float32 ...
utc_date (time) int32 1978012600 1978012601 ... 1978012622 1978012623
Attributes:
DATA_SOURCE: ECMWF: https://cds.climate.copernicus.eu...
NETCDF_CONVERSION: CISL RDA: Conversion from ECMWF GRIB 1 d...
NETCDF_VERSION: 4.8.1
CONVERSION_PLATFORM: Linux r1i3n32 4.12.14-95.51-default #1 S...
CONVERSION_DATE: Sun Jul 3 14:02:30 MDT 2022
Conventions: CF-1.6
NETCDF_COMPRESSION: NCO: Precision-preserving compression to...
history: Sun Jul 3 14:03:04 2022: ncks -4 --ppc ...
NCO: netCDF Operators version 5.0.3 (Homepage...
DODS_EXTRA.Unlimited_Dimension: timeCreate an Xarray DataArray object for Z
Z = dsZ['Z']
Z
<xarray.DataArray 'Z' (time: 24, level: 6, latitude: 721, longitude: 1440)>
[149506560 values with dtype=float32]
Coordinates:
* latitude (latitude) float64 90.0 89.75 89.5 89.25 ... -89.5 -89.75 -90.0
* level (level) float64 200.0 300.0 500.0 700.0 850.0 1e+03
* longitude (longitude) float64 0.0 0.25 0.5 0.75 ... 359.0 359.2 359.5 359.8
* time (time) datetime64[ns] 1978-01-26 ... 1978-01-26T23:00:00
Attributes:
long_name: Geopotential
short_name: z
units: m**2 s**-2
original_format: WMO GRIB 1 with ECMWF...
ecmwf_local_table: 128
ecmwf_parameter: 129
minimum_value: -4556.6367
maximum_value: 494008.56
grid_specification: 0.25 degree x 0.25 de...
rda_dataset: ds633.0
rda_dataset_url: https:/rda.ucar.edu/d...
rda_dataset_doi: DOI: 10.5065/BH6N-5N20
rda_dataset_group: ERA5 atmospheric pres...
QuantizeGranularBitGroomNumberOfSignificantDigits: 7
_ChunkSizes: [1, 37, 721, 1440]Set contour levels for both SLP and Z.
slpLevels = np.arange(864,1080,4)
Choose a particular isobaric surface.
pLevel = 500
Redefine Z so it consists only of the data at the level you just chose.
Z = Z.sel(level=pLevel)
Science-on-a-Sphere’s background maps require that projected images, besides using Plate Carree projection, extend from -180 to +180 degrees longitude. The ERA-5 datasets’ longitudinal coordinates start at 0 and go to 360. Although we won’t be using those maps (instead, we will add map borders to our graphic using Cartopy), let’s define a function that re-sorts datasets (or data arrays) so the longitudinal range begins at the dateline instead of the Greenwich meridian.
def reSortCoords(d):
longitudeName = 'longitude'
d.coords[longitudeName] = (d.coords[longitudeName] + 180) % 360 - 180
d = d.sortby(d[longitudeName])
return d
Z = reSortCoords(Z)
SLP = reSortCoords(SLP)
Examine one of the re-sorted data arrays
SLP
<xarray.DataArray 'MSL' (time: 24, latitude: 721, longitude: 1440)>
<Quantity([[[1032.5399 1032.5399 1032.5399 ... 1032.5399 1032.5399 1032.5399 ]
[1032.28 1032.2775 1032.2775 ... 1032.2825 1032.2825 1032.28 ]
[1031.955 1031.9525 1031.95 ... 1031.9625 1031.96 1031.9575 ]
...
[1008.3675 1008.375 1008.38495 ... 1008.33997 1008.35 1008.3575 ]
[1009.4975 1009.5025 1009.505 ... 1009.485 1009.4875 1009.4925 ]
[1009.69 1009.69 1009.69 ... 1009.69 1009.69 1009.69 ]]
[[1031.6238 1031.6238 1031.6238 ... 1031.6238 1031.6238 1031.6238 ]
[1031.3638 1031.3612 1031.3612 ... 1031.3662 1031.3662 1031.3638 ]
[1031.0387 1031.0363 1031.0337 ... 1031.0437 1031.0413 1031.0387 ]
...
[1008.32874 1008.33875 1008.34625 ... 1008.3012 1008.3087 1008.3187 ]
[1009.4912 1009.4962 1009.5012 ... 1009.47626 1009.4812 1009.4862 ]
[1009.70874 1009.70874 1009.70874 ... 1009.70874 1009.70874 1009.70874]]
[[1031.6018 1031.6018 1031.6018 ... 1031.6018 1031.6018 1031.6018 ]
[1031.3368 1031.3344 1031.3344 ... 1031.3394 1031.3394 1031.3368 ]
[1031.0194 1031.0168 1031.0144 ... 1031.0269 1031.0243 1031.0219 ]
...
...
...
[1005.88434 1005.89685 1005.91187 ... 1005.84186 1005.8569 1005.8693 ]
[1006.70935 1006.71686 1006.72186 ... 1006.68933 1006.69434 1006.70184]
[1006.89185 1006.89185 1006.89185 ... 1006.89185 1006.89185 1006.89185]]
[[1021.5062 1021.5062 1021.5062 ... 1021.5062 1021.5062 1021.5062 ]
[1021.08374 1021.08124 1021.07874 ... 1021.08875 1021.08624 1021.08624]
[1020.7362 1020.7337 1020.7287 ... 1020.7462 1020.7437 1020.7387 ]
...
[1008.1287 1008.13873 1008.14874 ... 1008.09875 1008.1087 1008.1187 ]
[1008.8537 1008.8587 1008.8637 ... 1008.83875 1008.84375 1008.84875]
[1008.97125 1008.97125 1008.97125 ... 1008.97125 1008.97125 1008.97125]]
[[1021.305 1021.305 1021.305 ... 1021.305 1021.305 1021.305 ]
[1020.86 1020.86 1020.8575 ... 1020.8675 1020.865 1020.8625 ]
[1020.3575 1020.3525 1020.3475 ... 1020.37 1020.365 1020.36 ]
...
[1007.8825 1007.89246 1007.90247 ... 1007.855 1007.865 1007.875 ]
[1008.67 1008.675 1008.68 ... 1008.65497 1008.66 1008.665 ]
[1008.8075 1008.8075 1008.8075 ... 1008.8075 1008.8075 1008.8075 ]]], 'hectopascal')>
Coordinates:
* latitude (latitude) float64 90.0 89.75 89.5 89.25 ... -89.5 -89.75 -90.0
* longitude (longitude) float64 -180.0 -179.8 -179.5 ... 179.2 179.5 179.8
* time (time) datetime64[ns] 1978-01-26 ... 1978-01-26T23:00:00
Attributes:
long_name: Mean sea level pressure
short_name: msl
original_format: WMO GRIB 1 with ECMWF...
ecmwf_local_table: 128
ecmwf_parameter: 151
minimum_value: 93279.19
maximum_value: 106472.31
grid_specification: 0.25 degree x 0.25 de...
rda_dataset: ds633.0
rda_dataset_url: https:/rda.ucar.edu/d...
rda_dataset_doi: DOI: 10.5065/BH6N-5N20
rda_dataset_group: ERA5 atmospheric surf...
QuantizeGranularBitGroomNumberOfSignificantDigits: 7
_ChunkSizes: [27, 139, 277]Notice that the longitude dimension now begins at -180.
For the purposes of plotting geopotential heights in decameters, choose an appropriate contour interval and range of values … for geopotential heights, a common convention is: from surface up through 700 hPa: 3 dam; above that, 6 dam to 400 and then 9 or 12 dam from 400 and above.
if (pLevel == 1000):
zLevels= np.arange(-90,63, 3)
elif (pLevel == 850):
zLevels = np.arange(60, 183, 3)
elif (pLevel == 700):
zLevels = np.arange(201, 339, 3)
elif (pLevel == 500):
zLevels = np.arange(468, 606, 6)
elif (pLevel == 300):
zLevels = np.arange(765, 1008, 9)
elif (pLevel == 200):
zLevels = np.arange(999, 1305, 9)
#print (U.units)
## Convert wind from m/s to knots; use MetPy to calculate the wind speed.
#UKts = U.metpy.convert_units('knots')
#VKts = V.metpy.convert_units('knots')
## Calculate windspeed.
#wspdKts = calc.wind_speed(U,V).to('knots')
Create objects for the relevant coordinate arrays; in this case, longitude, latitude, and time.
lons, lats, times= SLP.longitude, SLP.latitude, SLP.time
Take a peek at a couple of these coordinate arrays.
lons
<xarray.DataArray 'longitude' (longitude: 1440)> array([-180. , -179.75, -179.5 , ..., 179.25, 179.5 , 179.75]) Coordinates: * longitude (longitude) float64 -180.0 -179.8 -179.5 ... 179.2 179.5 179.8
add_cyclic_point method to eliminate the resulting seam.times
<xarray.DataArray 'time' (time: 24)>
array(['1978-01-26T00:00:00.000000000', '1978-01-26T01:00:00.000000000',
'1978-01-26T02:00:00.000000000', '1978-01-26T03:00:00.000000000',
'1978-01-26T04:00:00.000000000', '1978-01-26T05:00:00.000000000',
'1978-01-26T06:00:00.000000000', '1978-01-26T07:00:00.000000000',
'1978-01-26T08:00:00.000000000', '1978-01-26T09:00:00.000000000',
'1978-01-26T10:00:00.000000000', '1978-01-26T11:00:00.000000000',
'1978-01-26T12:00:00.000000000', '1978-01-26T13:00:00.000000000',
'1978-01-26T14:00:00.000000000', '1978-01-26T15:00:00.000000000',
'1978-01-26T16:00:00.000000000', '1978-01-26T17:00:00.000000000',
'1978-01-26T18:00:00.000000000', '1978-01-26T19:00:00.000000000',
'1978-01-26T20:00:00.000000000', '1978-01-26T21:00:00.000000000',
'1978-01-26T22:00:00.000000000', '1978-01-26T23:00:00.000000000'],
dtype='datetime64[ns]')
Coordinates:
* time (time) datetime64[ns] 1978-01-26 ... 1978-01-26T23:00:00
Attributes:
long_name: time
_ChunkSizes: 1024Create contour plots of SLP and geopotential height.¶
Let’s create a plot for a single time, just to ensure all looks good.
proj_data = ccrs.PlateCarree() # The dataset's x- and y- coordinates are lon-lat
Additionally, the sphere expects its graphics to have a resolution of 2048x1024.
Finally, by default, Matplotlib includes a border frame around each
Axes. We don't want that included on the sphere-projected graphic either.res = '110m'
dpi = 100
fig = plt.figure(figsize=(2048/dpi, 1024/dpi))
ax = plt.subplot(1,1,1,projection=ccrs.PlateCarree(), frameon=False)
ax.set_global()
ax.add_feature(cfeature.COASTLINE.with_scale(res))
ax.add_feature(cfeature.BORDERS.with_scale(res))
ax.add_feature(cfeature.STATES.with_scale(res))
# Convert from units of geopotential to height in decameters.
Zdam = mpcalc.geopotential_to_height(Z.isel(time=0)).metpy.convert_units('dam')
# add cyclic points to data and longitudes
# latitudes are unchanged in 1-dimension
SLP2d, clons= cutil.add_cyclic_point(SLP.isel(time=0), lons)
z2d, clons = cutil.add_cyclic_point(Zdam, lons)
# Height (Z) contour fills
# Note we don't need the transform argument since the map/data projections are the same, but we'll leave it in
CF = ax.contourf(clons,lats,z2d,levels=zLevels,cmap=plt.get_cmap('viridis'), extend='both', transform=proj_data)
# SLP contour lines
CL = ax.contour(clons,lats,SLP2d,slpLevels,linewidths=1.25,colors='chocolate', transform=proj_data)
ax.clabel(CL, inline_spacing=0.2, fontsize=8, fmt='%.0f')
fig.tight_layout(pad=.01)
# Save this figure to your current directory
fig.savefig('test_ERA_SOS.png')
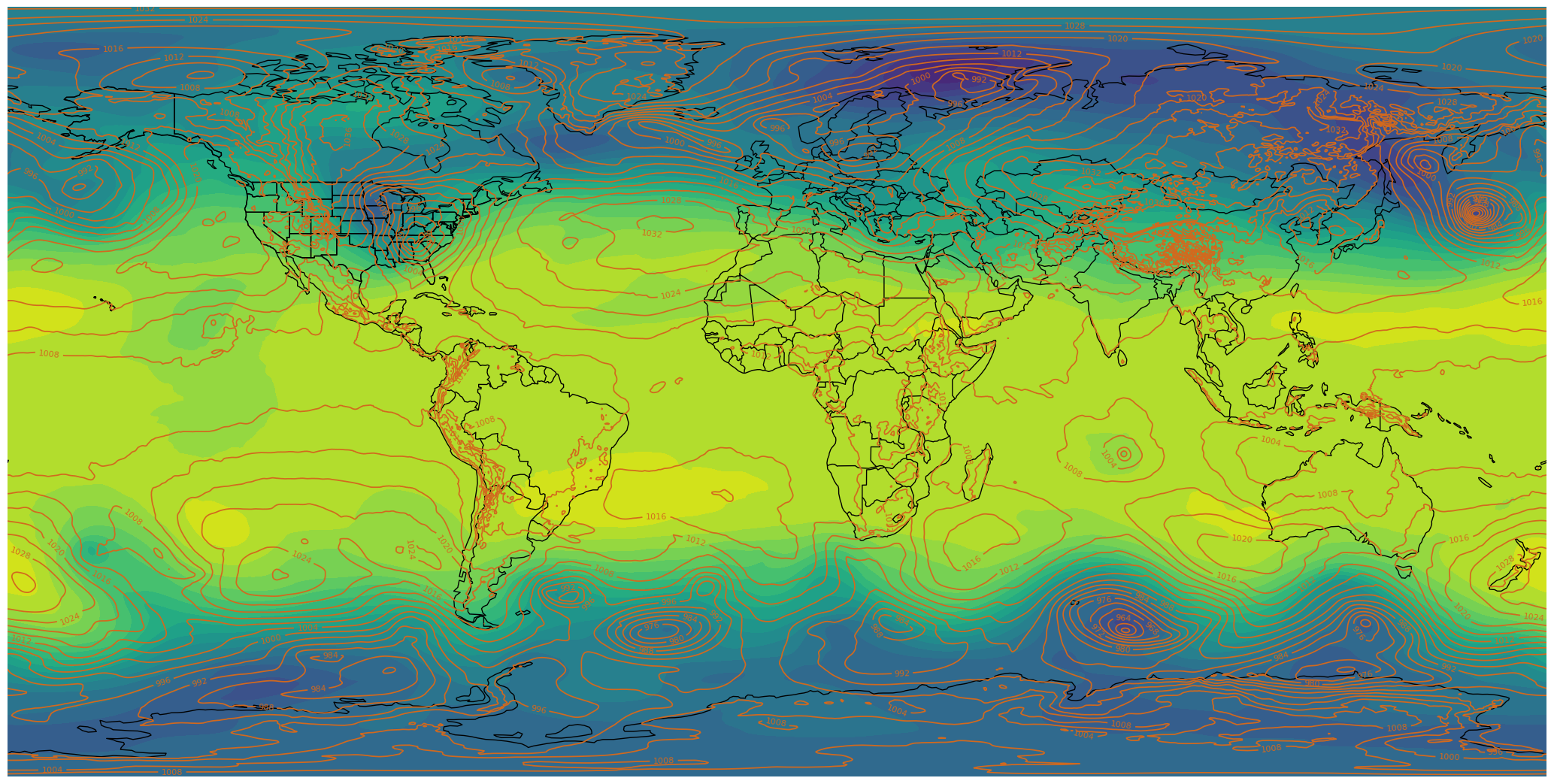
If you’d like, go to https://maptoglobe.com and view how your graphic might look on the sphere.¶
Follow these steps:
You should see the following after successfully connecting to the maptoglobe site:
Then, follow these steps:
Download the
test_ERA5_SOS.pngfile to your local computerClick on the Images tab on the maptoglobe site
Next to Surface, click on Choose a file
Navigate to the folder in which you downloaded the PNG, select the PNG, and click on OK
You should then see your map! Use your mouse to move the globe around; also use the mouse wheel to zoom in and out.
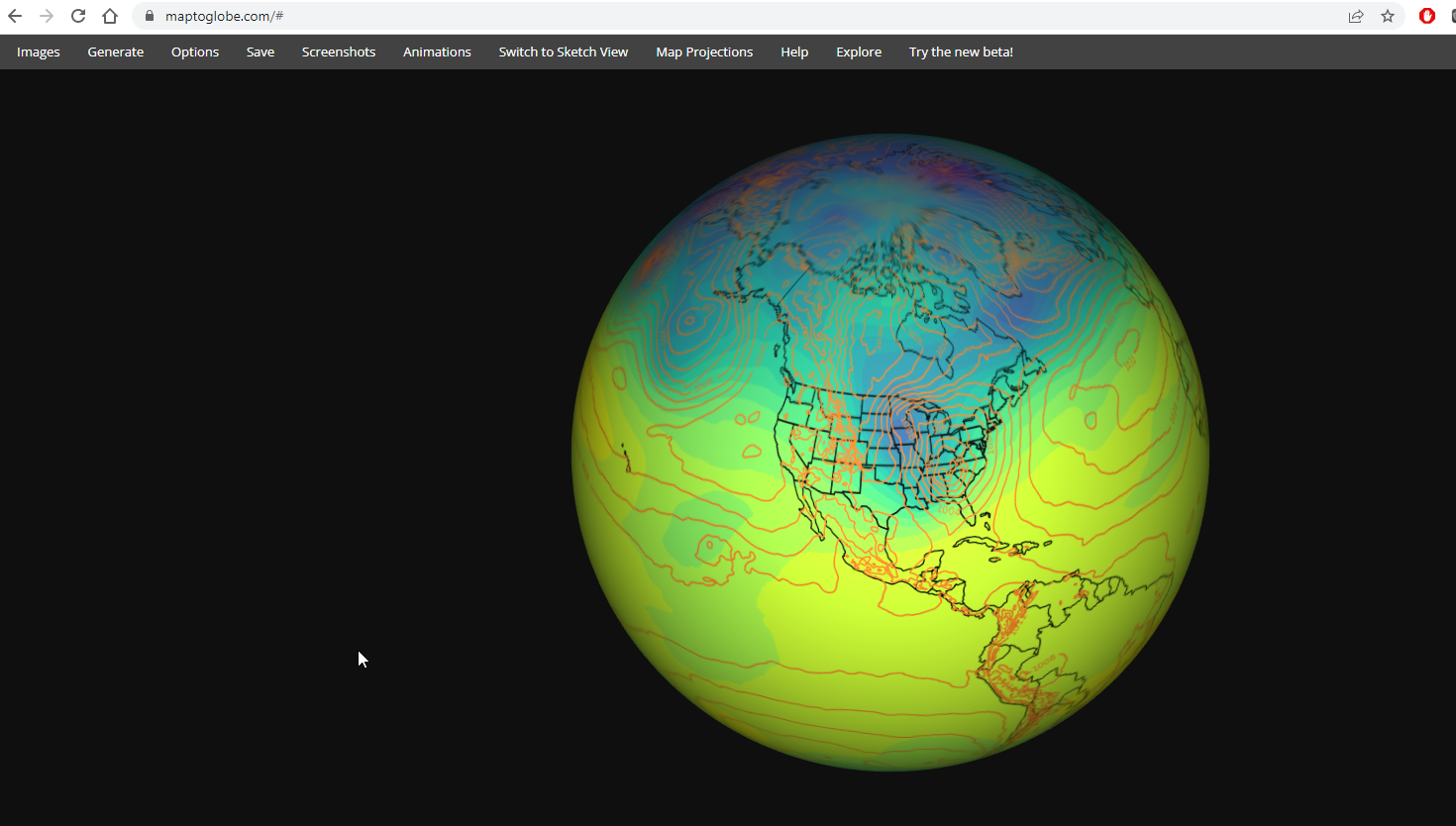
If the image looks good, you’re ready to move forward!
Create small (128x128) and large (800x800) thumbnails. These will serve as icons in the SoS iPad and computer apps that go along with your particular product.¶
We’ll use the orthographic projection and omit the contour lines and some of the cartographic features, and add a title string.
res = '110m'
dpi = 100
for size in (128, 800):
if (size == 128):
sizeStr = 'small'
else:
sizeStr ='big'
fig = plt.figure(figsize=(size/dpi, size/dpi))
ax = plt.subplot(1,1,1,projection=ccrs.Orthographic(central_longitude=-90), frameon=False)
ax.set_global()
ax.add_feature(cfeature.COASTLINE.with_scale(res))
tl1 = caseDir
tl2 = prodDir
ax.set_title(tl1 + '\n' + tl2, color='purple', fontsize=8)
# Height (Z) contour fills
CF = ax.contourf(clons,lats,z2d,levels=zLevels,cmap=plt.get_cmap('viridis'), extend='both', transform=proj_data)
fig.tight_layout(pad=.01)
fig.savefig(mediaDir + 'thumbnail_' + sizeStr + '.jpg')
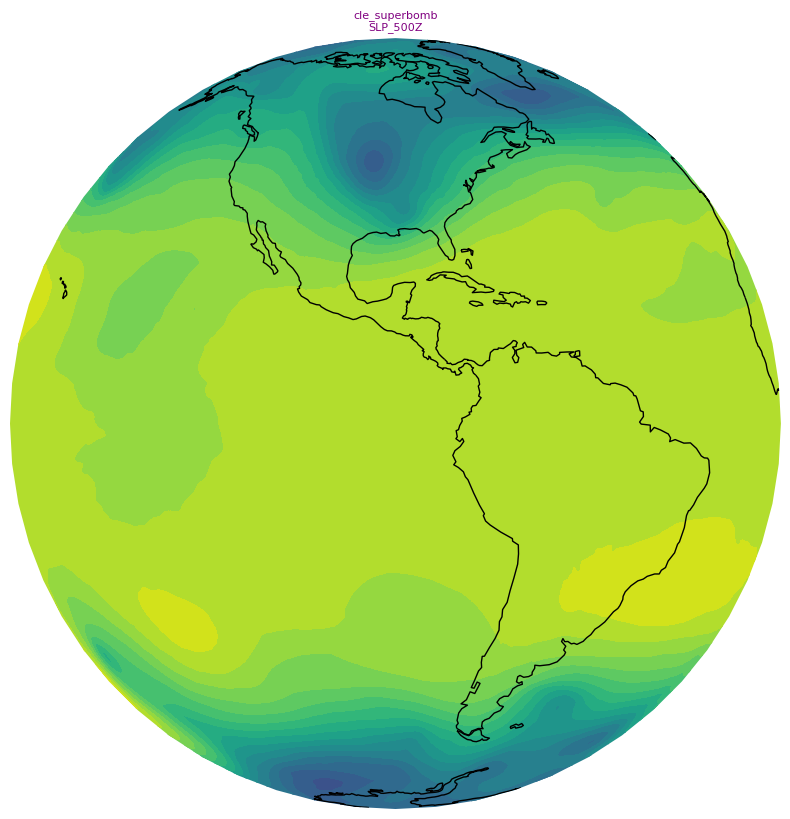
Create a standalone colorbar¶
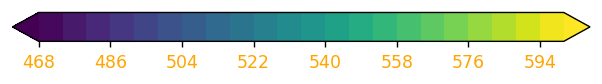
Visualizations on the Science-on-a-Sphere consist of a series of image files, layered on top of each other. In this example, instead of having the colorbar associated with 500 hPa heights adjacent to the map, let’s use a technique by which we remove the contour plot, leaving only the colorbar to be exported as an image. We change the colorbar’s orientation to horizontal, and also change its tick label colors so they will be more visible on the sphere’s display.
# draw a new figure and replot the colorbar there
fig,ax = plt.subplots(figsize=(14,2), dpi=125)
# set tick and ticklabel color
tick_color='black'
label_color='orange'
cbar = fig.colorbar(CF, ax=ax, orientation='horizontal')
cbar.ax.xaxis.set_tick_params(color=tick_color, labelcolor=label_color)
# Remove the Axes object ... essentially the contours and cartographic features ... from the figure.
ax.remove()
# All that remains is the colorbar ... save it to disk. Make the background transparent.
fig.savefig(labelsDir + 'colorbar.png',transparent=True)
Create a set of labels for each plot¶
labelFname = labelsDir + 'labels.txt'
Define a function to create the plot for each hour.¶
The function accepts the hour and a DataArray as its arguments.
def make_sos_map (i, z):
res = '110m'
dpi = 100
fig = plt.figure(figsize=(2048/dpi, 1024/dpi))
ax = plt.subplot(1,1,1,projection=ccrs.PlateCarree(), frameon=False)
ax.set_global()
ax.add_feature(cfeature.COASTLINE.with_scale(res))
ax.add_feature(cfeature.BORDERS.with_scale(res))
ax.add_feature(cfeature.STATES.with_scale(res))
# add cyclic points to data and longitudes
# latitudes are unchanged in 1-dimension
SLP2d, clons= cutil.add_cyclic_point(SLP.isel(time=i), lons)
z2d, clons = cutil.add_cyclic_point(z, lons)
# Height (Z) contour fills
CF = ax.contourf(clons,lats,z2d,levels=zLevels,cmap=plt.get_cmap('viridis'), extend='both')
# SLP contour lines
CL = ax.contour(clons,lats,SLP2d,slpLevels,linewidths=1.25,colors='chocolate', transform=proj_data)
ax.clabel(CL, inline_spacing=0.2, fontsize=8, fmt='%.0f')
fig.tight_layout(pad=.01)
frameNum = str(i).zfill(2)
figName = graphicsDir + "ERA5_"+ YYYYMMDD + frameNum + '.png'
fig.savefig(figName)
# Reduce the size of the PNG image via the Linux pngquant utility. The -f option overwites the resulting file if it already exists.
# The output file will end in "-fs8.png"
! pngquant -f {figName}
# Remove the original PNG
! rm -f {figName}
# Do not show the graphic in the notebook
plt.close()
Create the graphics and titles¶
Open a handle to the labels file
Define the time dimension index’s start, end, and increment values
Loop over the 24 hours
Perform any necessary unit conversions
Create each hour’s graphic
Write the title line to the text file.
Close the handle to the labels file
inc to 1 so all hours are processed. labelFileObject = open(labelFname, 'w')
startHr = 0
endHr = 24
inc = 12 # Change to 1 when ready
for time in range(startHr, endHr, inc):
# Perform any unit conversions appropriate for your scalar quantities. In this example,
# we convert geopotential to geopotential height in decameters.
z = mpcalc.geopotential_to_height(Z.isel(time=time)).metpy.convert_units('dam')
make_sos_map(time,z)
# Construct the title string and write it to the file
valTime = pd.to_datetime(times[time].values).strftime("%m/%d/%Y %H UTC")
tl1 = str("ERA-5 SLP & " + str(pLevel) + " hPa Z " + valTime + "\n") # \n is the newline character
labelFileObject.write(tl1)
print(tl1)
# Close the text file
labelFileObject.close()
ERA-5 SLP & 500 hPa Z 01/26/1978 00 UTC
ERA-5 SLP & 500 hPa Z 01/26/1978 12 UTC
Close the file handles to the remote RDA THREDDS server.¶
Modify as needed … close whatever Dataset objects you created via Xarray’s open_dataset method earlier in the notebook.
dsSLP.close()
dsZ.close()
Create an SoS playlist file¶
We follow the guidelines in https://sos.noaa.gov/support/sos/manuals/datasets/playlists/#dataset-playlist.sos-files
playlistFname = playlistDir + 'playlist.sos'
Open a file handle to the playlist file
Add each line of the playlist. Modify as needed for your case and product; in general you will only need to modify the name and description values!
Close the file handle
plFileObject = open(playlistFname, 'w')
cRet = "\n" # New line character code
nameStr = "Cleveland Superbomb 1978 SLP/500 Z"
descriptionStr = '{{ ERA-5 January 26 1978 ("Cleveland Superbomb") }}'
plFileObject.write("name = " + nameStr + cRet )
plFileObject.write("description = " + descriptionStr + cRet)
plFileObject.write("pip = ../labels/colorbar.png" + cRet)
plFileObject.write("pipheight = 12" + cRet)
plFileObject.write("pipvertical = -35" + cRet)
plFileObject.write("label = ../labels/labels.txt" + cRet)
plFileObject.write("layer = Grids" + cRet)
plFileObject.write("layerdata = ../2048" + cRet)
plFileObject.write("firstdwell = 2000" + cRet)
plFileObject.write("lastdwell = 3000" + cRet)
plFileObject.write("fps = 8" + cRet)
plFileObject.close()
Display the contents of the playlist file
! cat {playlistFname}
name = Cleveland Superbomb 1978 SLP/500 Z
description = {{ ERA-5 January 26 1978 ("Cleveland Superbomb") }}
pip = ../labels/colorbar.png
pipheight = 12
pipvertical = -35
label = ../labels/labels.txt
layer = Grids
layerdata = ../2048
firstdwell = 2000
lastdwell = 3000
fps = 8
- The name and description of the product, which are used in the SoS iPad and computer apps
- The three components of what is displayed on the screen:
- The colorbar (a picture-in-picture, aka pip), with its height and vertical position
- The label, or title, which describes the graphic
- The graphic layer itself. Multiple graphic layers, each with its own layer name and directory path, could be included; in this case, there is just one.
- The dwell times, in ms, for the first frame, last frame, and all frames in-between.