04_Xarray_Variables: More CFSR plotting
Contents

04_Xarray_Variables: More CFSR plotting¶
Overview¶
Select a date and access various CFSR Datasets
Subset the desired Datasets along their dimensions
Perform unit conversions
Create a well-labeled multi-parameter plot of gridded CFSR reanalysis data
Imports¶
import xarray as xr
import pandas as pd
import numpy as np
from datetime import datetime as dt
from metpy.units import units
import metpy.calc as mpcalc
import cartopy.crs as ccrs
import cartopy.feature as cfeature
import matplotlib.pyplot as plt
ERROR 1: PROJ: proj_create_from_database: Open of /knight/anaconda_aug22/envs/aug22_env/share/proj failed
Select a date and time, and access several CFSR Datasets¶
# Date/Time specification
Year = 1993
Month = 3
Day = 14
Hour = 12
Minute = 0
dateTime = dt(Year,Month,Day, Hour, Minute)
timeStr = dateTime.strftime("%Y-%m-%d %H%M UTC")
Create Xarray Dataset objects, each pointing to their respective NetCDF files in the /cfsr/data directory tree, using Xarray’s open_dataset method.¶
dsZ = xr.open_dataset (f'/cfsr/data/{Year}/g.{Year}.0p5.anl.nc')
dsT = xr.open_dataset (f'/cfsr/data/{Year}/t.{Year}.0p5.anl.nc')
dsU = xr.open_dataset (f'/cfsr/data/{Year}/u.{Year}.0p5.anl.nc')
dsV = xr.open_dataset (f'/cfsr/data/{Year}/v.{Year}.0p5.anl.nc')
dsQ = xr.open_dataset (f'/cfsr/data/{Year}/q.{Year}.0p5.anl.nc')
dsSLP = xr.open_dataset (f'/cfsr/data/{Year}/pmsl.{Year}.0p5.anl.nc')
Let’s examine one of the Datasets.¶
dsZ
<xarray.Dataset>
Dimensions: (time: 1460, lat: 361, lon: 720, lev: 32)
Coordinates:
* time (time) datetime64[ns] 1993-01-01 ... 1993-12-31T18:00:00
* lat (lat) float32 -90.0 -89.5 -89.0 -88.5 -88.0 ... 88.5 89.0 89.5 90.0
* lon (lon) float32 -180.0 -179.5 -179.0 -178.5 ... 178.5 179.0 179.5
* lev (lev) float32 1e+03 975.0 950.0 925.0 900.0 ... 50.0 30.0 20.0 10.0
Data variables:
g (time, lev, lat, lon) float32 ...
Attributes:
description: g 1000-10 hPa
year: 1993
source: http://nomads.ncdc.noaa.gov/data.php?name=access#CFSR-data
references: Saha, et. al., (2010)
created_by: User: kgriffin
creation_date: Wed Apr 4 05:58:31 UTC 2012Subset the desired Datasets along their dimensions.¶
We will use the sel method on the latitude and longitude dimensions as well as time and isobaric surface.¶
# Areal extent
lonW = -100.
lonE = -60.
latS = 20.
latN = 50.
cLat, cLon = (latS + latN)/2, (lonW + lonE)/2
expand = 0.5
latRange = np.arange(latS,latN + expand, .5) # expand the data range a bit beyond the plot range
lonRange = np.arange(lonW,lonE + expand, .5) # Need to match longitude values to those of the coordinate variable
# Vertical level specificaton
plevel = 850
levelStr = f'{plevel}' # Use for the figure title
# Data variable selection; modify depending on what variables you are interested in
T = dsT['t'].sel(time=dateTime,lev=plevel,lat=latRange,lon=lonRange)
U = dsU['u'].sel(time=dateTime,lev=plevel,lat=latRange,lon=lonRange)
V = dsV['v'].sel(time=dateTime,lev=plevel,lat=latRange,lon=lonRange)
Output the level and time strings; we’ll use them to create the figure’s title.¶
levelStr, timeStr
('850', '1993-03-14 1200 UTC')
Perform unit conversions¶
We take the DataArrays and apply MetPy’s unit conversion method.¶
T = T.metpy.convert_units('degC')
U = U.metpy.convert_units('kts')
V = V.metpy.convert_units('kts')
Create a well-labeled multi-parameter plot of gridded CFSR reanalysis data¶
proj_map = ccrs.LambertConformal(central_longitude=cLon, central_latitude=cLat)
proj_data = ccrs.PlateCarree() # Our data is lat-lon; thus its native projection is Plate Carree.
res = '50m'
Now define the range of our contour values and a contour interval.¶
This of course will depend on the level, time, region, and variable.
T.min(), T.max()
(<xarray.DataArray 't' ()>
<Quantity(-26.749984741210938, 'degree_Celsius')>
Coordinates:
time datetime64[ns] 1993-03-14T12:00:00
lev float32 850.0,
<xarray.DataArray 't' ()>
<Quantity(16.95001220703125, 'degree_Celsius')>
Coordinates:
time datetime64[ns] 1993-03-14T12:00:00
lev float32 850.0)
minTVal = -30
maxTVal = 22
cint = 2
Tcintervals = np.arange(minTVal, maxTVal, cint)
Tcintervals
array([-30, -28, -26, -24, -22, -20, -18, -16, -14, -12, -10, -8, -6,
-4, -2, 0, 2, 4, 6, 8, 10, 12, 14, 16, 18, 20])
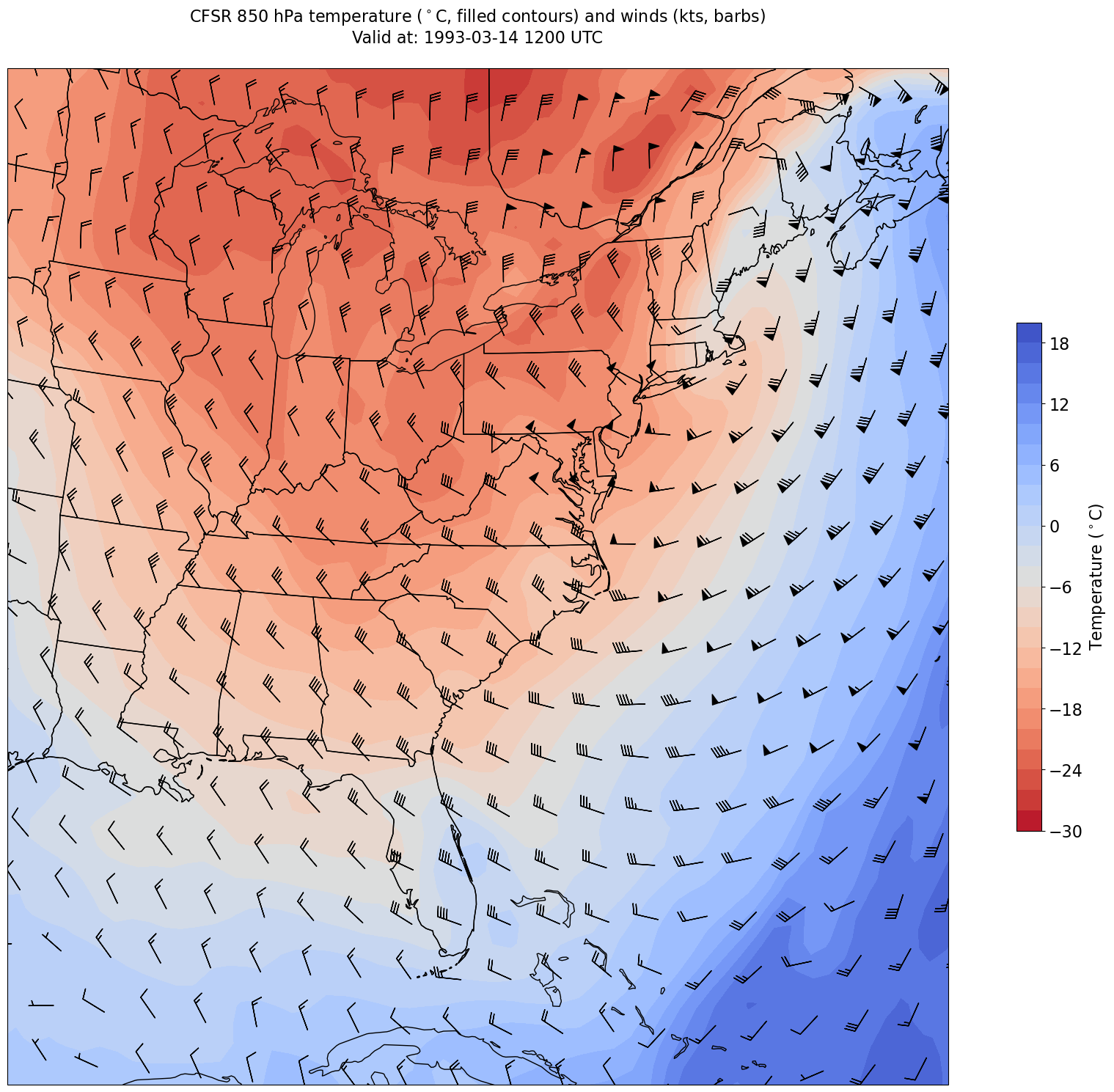
Plot the map¶
In this example: filled contours of 850 hPa temperature and wind barbs.
Create a meaningful title string.
tl1 = f'CFSR {levelStr} hPa temperature ($^\circ$C, filled contours) and winds (kts, barbs)'
tl2 = f'Valid at: {timeStr}'
title_line = (tl1 + '\n' + tl2 + '\n') # concatenate the two strings and add return characters
constrainLat, constrainLon = (0.7, 6.5)
fig = plt.figure(figsize=(24,18)) # Increase size to adjust for the constrained lats/lons
ax = plt.subplot(1,1,1,projection=proj_map)
ax.set_extent ([lonW+constrainLon,lonE-constrainLon,latS+constrainLat,latN-constrainLat])
ax.add_feature(cfeature.COASTLINE.with_scale(res))
ax.add_feature(cfeature.STATES.with_scale(res))
CF = ax.contourf(lons,lats,T, levels=Tcintervals,transform=proj_data,cmap='coolwarm_r')
cbar = plt.colorbar(CF,shrink=0.5)
cbar.ax.tick_params(labelsize=16)
cbar.ax.set_ylabel("Temperature ($^\circ$C)",fontsize=16)
# Plotting wind barbs uses the ax.barbs method. Here, you can't pass in the DataArray directly; you can only pass in the array's values.
# Also need to sample (skip) a selected # of points to keep the plot readable.
skip = 3
ax.barbs(lons[::skip],lats[::skip],U[::skip,::skip].values, V[::skip,::skip].values, transform=proj_data)
title = plt.title(title_line,fontsize=16)